You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_435205-t37_1-p1
Basic Information
help
| Species |
Aspergillus novofumigatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus
|
| CAZyme ID |
P174DRAFT_435205-t37_1-p1
|
| CAZy Family |
GH18 |
| CAZyme Description |
pectin lyase-like protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 486 |
|
53464.45 |
6.7327 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AnovofumigatusIBT16806 |
11729 |
1392255 |
197 |
11532
|
|
| Gene Location |
No EC number prediction in P174DRAFT_435205-t37_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH28 |
226 |
453 |
1e-32 |
0.6430769230769231 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 215426
|
PLN02793 |
0.001 |
227 |
347 |
230 |
359 |
Probable polygalacturonase |
| 227721
|
Pgu1 |
0.004 |
190 |
301 |
216 |
319 |
Polygalacturonase [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 8.64e-232 |
4 |
486 |
5 |
490 |
| 2.30e-192 |
24 |
484 |
15 |
483 |
| 2.65e-192 |
24 |
484 |
19 |
487 |
| 2.65e-192 |
24 |
484 |
19 |
487 |
| 9.22e-177 |
24 |
472 |
19 |
475 |
P174DRAFT_435205-t37_1-p1 has no PDB hit.
P174DRAFT_435205-t37_1-p1 has no Swissprot hit.
This protein is predicted as SP
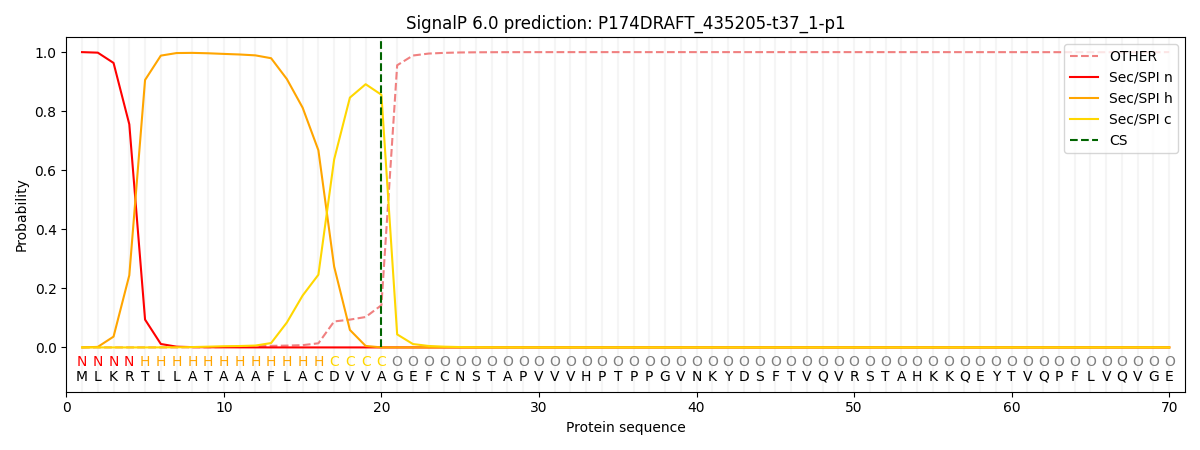
| Other |
SP_Sec_SPI |
CS Position |
| 0.000890 |
0.999079 |
CS pos: 20-21. Pr: 0.8555 |
There is no transmembrane helices in P174DRAFT_435205-t37_1-p1.

