You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_430314-t37_1-p1
Basic Information
help
| Species |
Aspergillus novofumigatus
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus
|
| CAZyme ID |
P174DRAFT_430314-t37_1-p1
|
| CAZy Family |
PL4 |
| CAZyme Description |
glycoside hydrolase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 456 |
MSZS01000003|CGC15 |
50514.56 |
9.0692 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_AnovofumigatusIBT16806 |
11729 |
1392255 |
197 |
11532
|
|
| Gene Location |
No EC number prediction in P174DRAFT_430314-t37_1-p1.
P174DRAFT_430314-t37_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 5.43e-228 |
1 |
453 |
1 |
460 |
| 1.07e-189 |
8 |
455 |
7 |
442 |
| 1.07e-189 |
8 |
455 |
7 |
442 |
| 8.13e-180 |
26 |
453 |
18 |
432 |
| 5.72e-133 |
1 |
455 |
1 |
445 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.78e-14 |
154 |
375 |
119 |
324 |
Structure of N2152 from Neocallimastix frontalis [Neocallimastix frontalis] |
P174DRAFT_430314-t37_1-p1 has no Swissprot hit.
This protein is predicted as SP
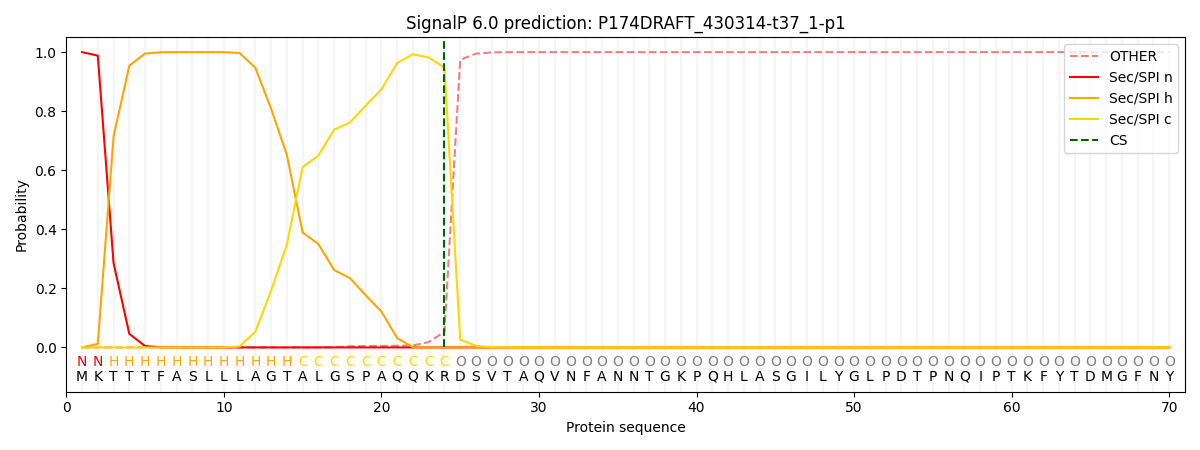
| Other |
SP_Sec_SPI |
CS Position |
| 0.000577 |
0.999388 |
CS pos: 24-25. Pr: 0.9473 |
There is no transmembrane helices in P174DRAFT_430314-t37_1-p1.

