You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_425759-t37_1-p1
You are here: Home > Sequence: P174DRAFT_425759-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_425759-t37_1-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | fibronectin type III domain protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH135 | 473 | 704 | 2.6e-21 | 0.9493670886075949 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403383 | Spherulin4 | 5.05e-48 | 476 | 712 | 1 | 238 | Spherulation-specific family 4. This protein is found in bacteria, archaea and eukaryotes. Proteins in this family are typically between 250 and 398 amino acids in length. There is a conserved NPG sequence motif and there are two completely conserved G residues that may be functionally important. Starvation will often induce spherulation - the production of spores - and this process may involve DNA-methylation. Changes in the methylation of spherulin4 are associated with the formation of spherules, but these changes are probably transient. Methylation of the gene accompanies its transcriptional activation, and spherulin4 mRNA is only detectable in late spherulating cultures and mature spherules. It is a spherulation-specific protein. |
| 238861 | SEST_like | 2.31e-34 | 58 | 310 | 2 | 224 | SEST_like. A family of secreted SGNH-hydrolases similar to Streptomyces scabies esterase (SEST), a causal agent of the potato scab disease, which hydrolyzes a specific ester bond in suberin, a plant lipid. The tertiary fold of this enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles two of the three components of typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxylic acid. |
| 406036 | MU117 | 1.87e-30 | 981 | 1088 | 1 | 104 | Meiotically up-regulated gene family. This protein was identified as being up-regulated during meiosis in S.pombe. This family of proteins is found in largely in plants and fungi. Proteins in this family are typically between 128 and 920 amino acids in length. |
| 404371 | Lipase_GDSL_2 | 1.57e-05 | 61 | 146 | 1 | 74 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 400568 | ChitinaseA_N | 3.55e-04 | 743 | 832 | 30 | 130 | Chitinase A, N-terminal domain. This domain is found in a number of bacterial chitinases and similar viral proteins. It is organized into a fibronectin III module domain-like fold, comprising only beta strands. Its function is not known, but it may be involved in interaction with the enzyme substrate, chitin. It is separated by a hinge region from the catalytic domain (pfam00704); this hinge region is probably mobile, allowing the N-terminal domain to have different relative positions in solution. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1095 | 1 | 1114 | |
| 0.0 | 1 | 1095 | 1 | 1114 | |
| 3.13e-139 | 458 | 974 | 486 | 1000 | |
| 1.21e-120 | 4 | 463 | 3 | 463 | |
| 1.23e-120 | 4 | 463 | 3 | 463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.73e-15 | 61 | 280 | 7 | 176 | Crystal structure of phospholipase A1 from Streptomyces albidoflavus NA297 [Streptomyces albidoflavus] |
|
| 3.15e-12 | 57 | 281 | 2 | 179 | Crystal structure of extracelular lipase from Streptomyces rimosus at 1.7A resolution [Streptomyces rimosus],5MAL_B Crystal structure of extracelular lipase from Streptomyces rimosus at 1.7A resolution [Streptomyces rimosus] |
|
| 5.92e-07 | 466 | 675 | 5 | 211 | Crystal Structure of Aspergillus clavatus Sph3 [Aspergillus clavatus NRRL 1] |
|
| 6.96e-07 | 466 | 675 | 23 | 229 | Crystal Structure of Aspergillus clavatus Sph3 in complex with GalNAc [Aspergillus clavatus NRRL 1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.63e-13 | 61 | 282 | 40 | 213 | Lipase 1 OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO1725 PE=1 SV=1 |
|
| 2.71e-11 | 57 | 281 | 36 | 213 | Lipase OS=Streptomyces rimosus OX=1927 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
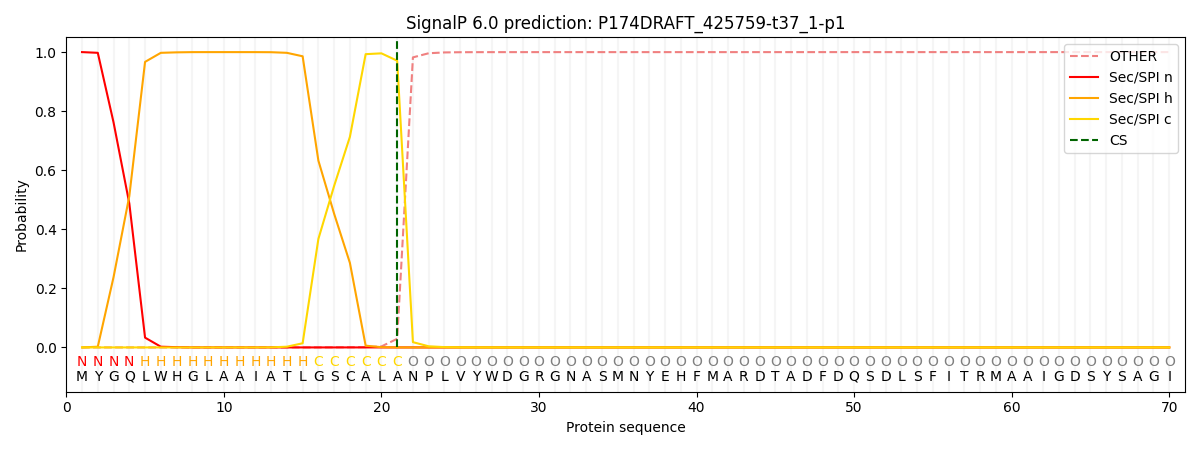
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000291 | 0.999669 | CS pos: 21-22. Pr: 0.9709 |

