You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_425181-t37_1-p1
You are here: Home > Sequence: P174DRAFT_425181-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_425181-t37_1-p1 | |||||||||||
| CAZy Family | GH17 | |||||||||||
| CAZyme Description | alginate lyase 2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL7 | 34 | 247 | 1.2e-83 | 0.9953271028037384 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400923 | Alginate_lyase2 | 2.34e-66 | 34 | 247 | 1 | 222 | Alginate lyase. Alginate lyases are enzymes that degrade the linear polysaccharide alignate. They cleave the glycosidic linkage of alignate through a beta-elimination reaction. This family forms an all beta fold and is different to all alpha fold of pfam05426. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.04e-101 | 17 | 247 | 18 | 250 | |
| 2.80e-96 | 23 | 247 | 19 | 249 | |
| 3.85e-96 | 13 | 247 | 14 | 247 | |
| 3.06e-93 | 23 | 247 | 19 | 249 | |
| 5.95e-93 | 23 | 247 | 18 | 248 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.29e-58 | 27 | 247 | 6 | 231 | Chain A, Glucuronan lyase [Trichoderma parareesei] |
|
| 1.12e-20 | 27 | 247 | 2 | 226 | Chain A, Alginate lyase [unidentified] |
|
| 1.60e-20 | 27 | 247 | 3 | 227 | Crystal structure at 1.0 A of alginate lyase A1-II', a member of polysaccharide lyase family-7 [Sphingomonas sp. A1],2Z42_A Crystal Structure of Family 7 Alginate Lyase A1-II' from Sphingomonas sp. A1 [Sphingomonas sp.] |
|
| 5.87e-20 | 27 | 247 | 2 | 226 | Chain A, Alginate lyase [Sphingomonas sp. A1],2ZAC_A Chain A, Alginate lyase [Sphingomonas sp. A1] |
|
| 5.90e-19 | 27 | 247 | 2 | 226 | Chain A, Alginate lyase [Sphingomonas sp. A1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.78e-07 | 23 | 231 | 18 | 282 | Alginate lyase OS=Klebsiella pneumoniae OX=573 GN=alyA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
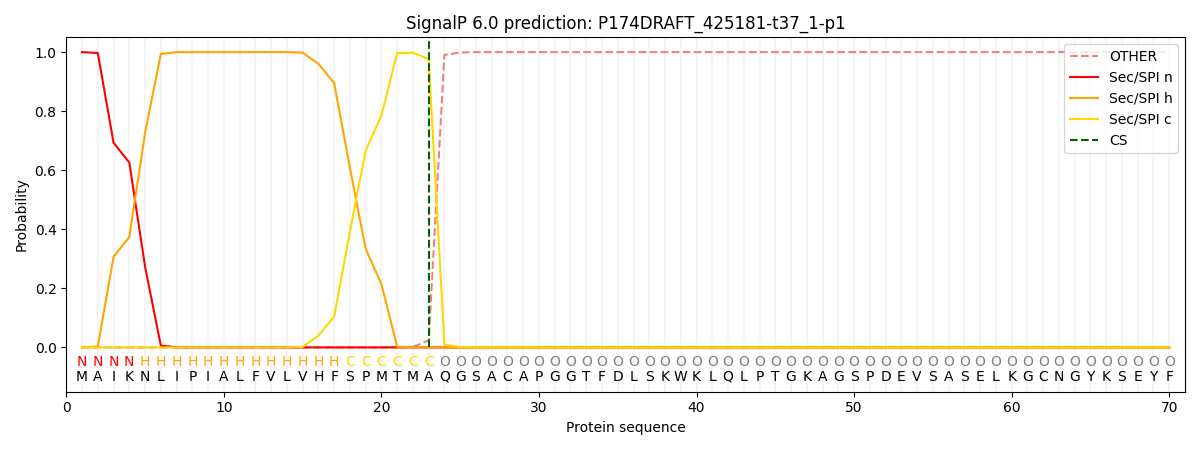
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000193 | 0.999778 | CS pos: 23-24. Pr: 0.9764 |
