You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_409639-t37_1-p1
You are here: Home > Sequence: P174DRAFT_409639-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_409639-t37_1-p1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | putative cutinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.74:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 38 | 214 | 3e-46 | 0.9894179894179894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 5.63e-60 | 38 | 215 | 1 | 173 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.69e-101 | 1 | 215 | 1 | 219 | |
| 7.69e-101 | 1 | 215 | 1 | 219 | |
| 6.84e-97 | 3 | 216 | 4 | 225 | |
| 6.84e-97 | 3 | 216 | 4 | 225 | |
| 1.21e-96 | 1 | 215 | 3 | 225 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.20e-69 | 23 | 215 | 5 | 198 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 6.46e-69 | 23 | 215 | 5 | 196 | Crystal structure of Aspergillus oryzae cutinase [Aspergillus oryzae] |
|
| 8.80e-67 | 32 | 215 | 5 | 187 | Structure of Aspergillus oryzae cutinase expressed in Pichia pastoris, crystallized in the presence of Paraoxon [Aspergillus oryzae] |
|
| 5.51e-62 | 28 | 216 | 3 | 193 | Humicola insolens cutinase [Humicola insolens],4OYY_B Humicola insolens cutinase [Humicola insolens],4OYY_C Humicola insolens cutinase [Humicola insolens],4OYY_D Humicola insolens cutinase [Humicola insolens],4OYY_E Humicola insolens cutinase [Humicola insolens],4OYY_F Humicola insolens cutinase [Humicola insolens],4OYY_G Humicola insolens cutinase [Humicola insolens],4OYY_H Humicola insolens cutinase [Humicola insolens],4OYY_I Humicola insolens cutinase [Humicola insolens],4OYY_J Humicola insolens cutinase [Humicola insolens],4OYY_K Humicola insolens cutinase [Humicola insolens],4OYY_L Humicola insolens cutinase [Humicola insolens] |
|
| 2.08e-61 | 19 | 216 | 6 | 208 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.82e-151 | 1 | 217 | 1 | 217 | Probable cutinase 3 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_030250 PE=3 SV=1 |
|
| 8.54e-141 | 1 | 216 | 1 | 216 | Probable cutinase 3 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_4G03210 PE=3 SV=1 |
|
| 8.54e-141 | 1 | 216 | 1 | 216 | Probable cutinase 3 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_099910 PE=3 SV=1 |
|
| 1.15e-117 | 1 | 215 | 1 | 215 | Probable cutinase 3 OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=ACLA_055320 PE=3 SV=1 |
|
| 1.50e-109 | 1 | 216 | 1 | 218 | Probable cutinase 3 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=ATEG_04791 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
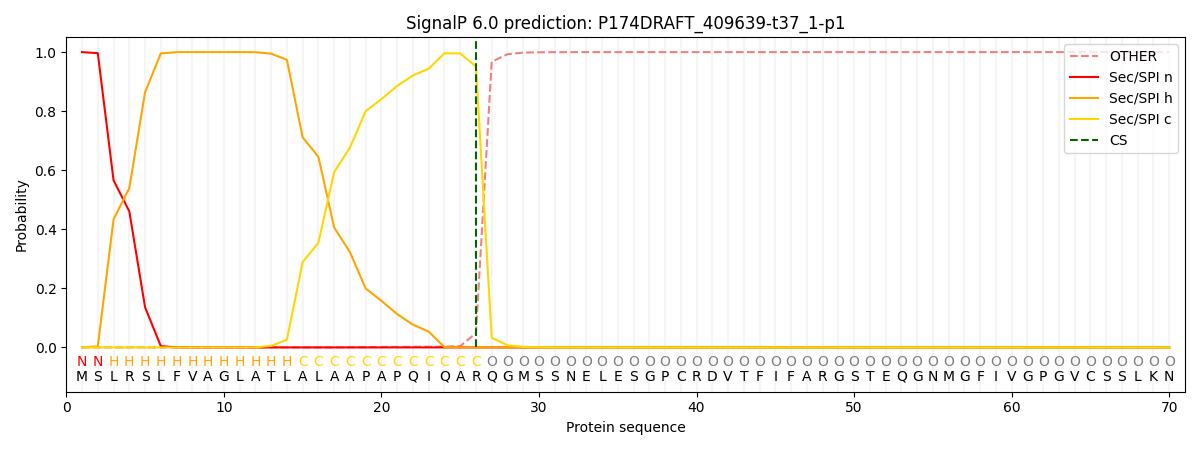
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000272 | 0.999701 | CS pos: 26-27. Pr: 0.9515 |
