You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_403605-t37_1-p1
You are here: Home > Sequence: P174DRAFT_403605-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_403605-t37_1-p1 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | putative cell wall glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 105 | 361 | 1.6e-23 | 0.9646302250803859 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 6.47e-68 | 90 | 357 | 30 | 300 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 0.004 | 291 | 357 | 230 | 296 | Glycosyl hydrolases family 17. |
| 381626 | GH113_mannanase-like | 0.005 | 151 | 331 | 70 | 273 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 7.37e-238 | 22 | 371 | 7 | 375 | |
| 1.64e-137 | 7 | 357 | 8 | 360 | |
| 1.64e-137 | 7 | 357 | 8 | 360 | |
| 9.43e-137 | 2 | 358 | 3 | 361 | |
| 1.09e-118 | 1 | 357 | 1 | 362 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.62e-67 | 105 | 357 | 128 | 375 | Cell surface mannoprotein MP65 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MP65 PE=1 SV=3 |
|
| 1.38e-64 | 105 | 357 | 135 | 382 | Probable family 17 glucosidase SCW4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW4 PE=1 SV=1 |
|
| 4.17e-64 | 105 | 357 | 138 | 385 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 1.12e-40 | 100 | 360 | 305 | 563 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 1.12e-40 | 100 | 360 | 305 | 563 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
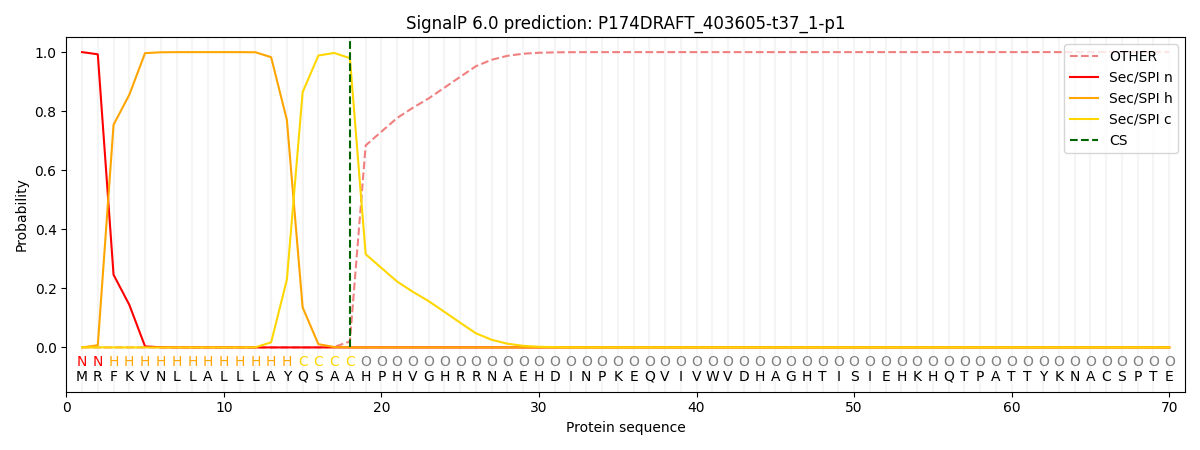
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000236 | 0.999715 | CS pos: 18-19. Pr: 0.9795 |
