You are browsing environment: FUNGIDB
CAZyme Information: P174DRAFT_129096-t37_1-p1
You are here: Home > Sequence: P174DRAFT_129096-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus novofumigatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus novofumigatus | |||||||||||
| CAZyme ID | P174DRAFT_129096-t37_1-p1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 18 | 216 | 1.7e-67 | 0.9633507853403142 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223021 | PHA03247 | 7.79e-10 | 286 | 695 | 2580 | 2984 | large tegument protein UL36; Provisional |
| 223021 | PHA03247 | 7.85e-08 | 267 | 670 | 2597 | 2999 | large tegument protein UL36; Provisional |
| 411414 | MSCRAMM_ClfB | 2.01e-06 | 340 | 626 | 554 | 833 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| 411414 | MSCRAMM_ClfB | 2.97e-06 | 368 | 662 | 548 | 841 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
| 411414 | MSCRAMM_ClfB | 3.07e-06 | 486 | 734 | 546 | 784 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.44e-99 | 1 | 244 | 1 | 238 | |
| 1.44e-99 | 1 | 244 | 1 | 238 | |
| 1.44e-99 | 1 | 244 | 1 | 238 | |
| 1.44e-99 | 1 | 244 | 1 | 238 | |
| 1.44e-99 | 1 | 244 | 1 | 238 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.51e-31 | 18 | 215 | 1 | 200 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
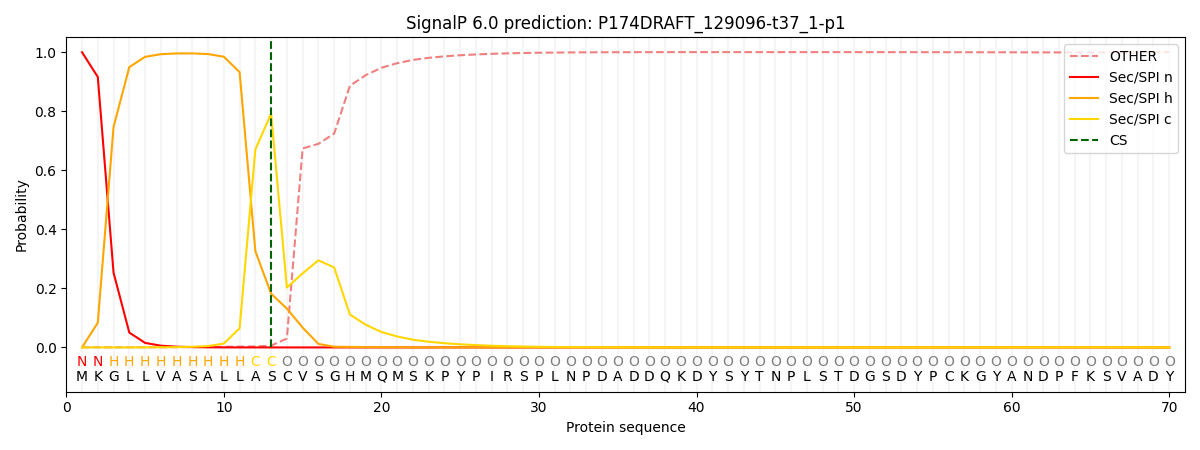
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000969 | 0.999027 | CS pos: 13-14. Pr: 0.7919 |
