You are browsing environment: FUNGIDB
CAZyme Information: P170DRAFT_462269-t37_1-p1
You are here: Home > Sequence: P170DRAFT_462269-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus steynii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus steynii | |||||||||||
| CAZyme ID | P170DRAFT_462269-t37_1-p1 | |||||||||||
| CAZy Family | GH78 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.259:12 | 2.4.1.261:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 39 | 463 | 1.8e-95 | 0.9948586118251928 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 403214 | DUF3445 | 1.66e-102 | 787 | 1021 | 1 | 230 | Protein of unknown function (DUF3445). This family of proteins are functionally uncharacterized. This protein is found in bacteria and eukaryotes. Proteins in this family are typically between 264 to 418 amino acids in length. This protein has a conserved RLP sequence motif. This protein has two completely conserved R residues that may be functionally important. |
| 281842 | Glyco_transf_22 | 4.19e-85 | 37 | 463 | 1 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 224720 | ArnT | 3.70e-04 | 39 | 378 | 14 | 340 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| 215437 | PLN02816 | 0.008 | 49 | 375 | 53 | 358 | mannosyltransferase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 5 | 1082 | 3 | 960 | |
| 0.0 | 29 | 1087 | 14 | 1112 | |
| 0.0 | 5 | 599 | 3 | 597 | |
| 0.0 | 1 | 599 | 1 | 597 | |
| 0.0 | 1 | 599 | 1 | 597 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.56e-17 | 791 | 1064 | 58 | 333 | Crystal structure of the alpha subunit of heme dependent oxidative N-demethylase (HODM) [Pseudomonas mendocina ymp],5LTH_A Crystal structure of the alpha subunit of heme dependent oxidative N-demethylase (HODM) in complex with the dimethylamine substrate [Pseudomonas mendocina],5LTI_A Crystal structure of the alpha subunit of heme dependent oxidative N-demethylase (HODM) in complex with the dimethylamine substrate [Pseudomonas mendocina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.76e-118 | 38 | 593 | 17 | 560 | Alpha-1,2-mannosyltransferase alg9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg9 PE=3 SV=1 |
|
| 1.04e-97 | 32 | 593 | 54 | 600 | Alpha-1,2-mannosyltransferase ALG9 OS=Mus musculus OX=10090 GN=Alg9 PE=2 SV=1 |
|
| 2.71e-95 | 32 | 605 | 54 | 610 | Alpha-1,2-mannosyltransferase ALG9 OS=Homo sapiens OX=9606 GN=ALG9 PE=1 SV=2 |
|
| 1.23e-84 | 33 | 597 | 6 | 554 | Alpha-1,2-mannosyltransferase ALG9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG9 PE=1 SV=1 |
|
| 1.99e-77 | 38 | 573 | 55 | 585 | Alpha-1,2-mannosyltransferase algn-9 OS=Caenorhabditis elegans OX=6239 GN=algn-9 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
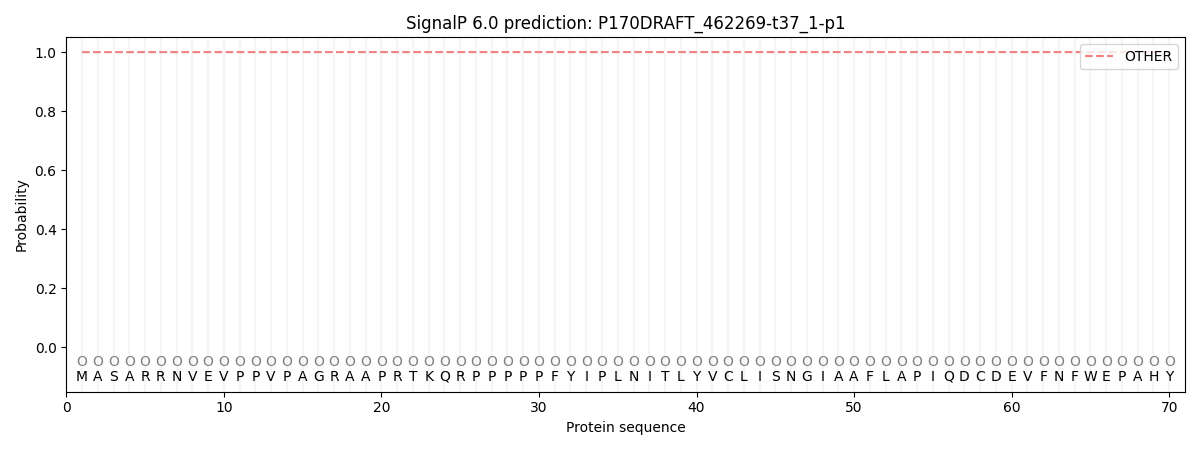
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999983 | 0.000020 |
TMHMM Annotations download full data without filtering help
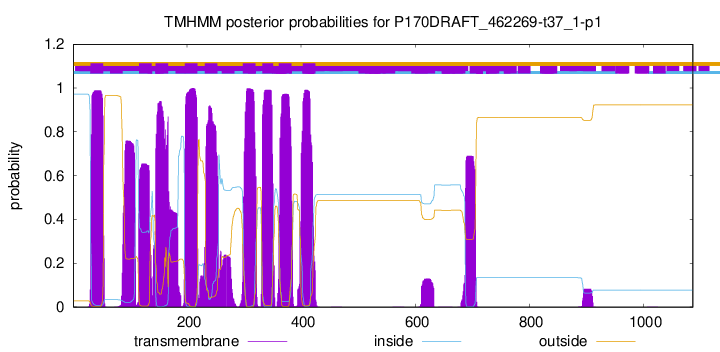
| Start | End |
|---|---|
| 31 | 53 |
| 116 | 138 |
| 145 | 167 |
| 196 | 218 |
| 231 | 253 |
| 298 | 320 |
| 332 | 351 |
| 361 | 383 |
| 403 | 425 |
