You are browsing environment: FUNGIDB
CAZyme Information: P170DRAFT_444217-t37_1-p1
You are here: Home > Sequence: P170DRAFT_444217-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus steynii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus steynii | |||||||||||
| CAZyme ID | P170DRAFT_444217-t37_1-p1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | endoglucanase B | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.-:12 | 3.2.1.4:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 54 | 321 | 1.6e-68 | 0.9891304347826086 |
| CBM46 | 366 | 447 | 2.9e-16 | 0.9080459770114943 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395098 | Cellulase | 2.40e-34 | 54 | 321 | 16 | 269 | Cellulase (glycosyl hydrolase family 5). |
| 225344 | BglC | 9.43e-19 | 34 | 321 | 47 | 360 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 397483 | CBM_X2 | 2.00e-18 | 369 | 453 | 6 | 82 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.95e-313 | 21 | 571 | 24 | 574 | |
| 4.68e-309 | 2 | 571 | 3 | 572 | |
| 1.36e-299 | 1 | 571 | 1 | 574 | |
| 1.18e-295 | 22 | 571 | 44 | 592 | |
| 1.43e-288 | 1 | 571 | 1 | 569 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.01e-95 | 27 | 557 | 20 | 524 | Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis [Bacillus licheniformis],4YZT_A Crystal structure of a tri-modular GH5 (subfamily 4) endo-beta-1, 4-glucanase from Bacillus licheniformis complexed with cellotetraose [Bacillus licheniformis] |
|
| 9.02e-58 | 34 | 507 | 13 | 477 | Structural Insight of a Trimodular Halophilic Cellulase with a Family 46 Carbohydrate-Binding Module [Bacillus sp. BG-CS10] |
|
| 2.26e-57 | 34 | 507 | 46 | 510 | A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10],5XRC_B A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10],5XRC_C A Trimodular GH5_4 Subfamily Endoglucanase Structure with Large Unit Cell [Bacillus sp. BG-CS10] |
|
| 1.73e-56 | 34 | 507 | 13 | 477 | Structural Insight of a Trimodular Halophilic Cellulase with a Family 46 Carbohydrate-Binding Module [Bacillus sp. BG-CS10] |
|
| 8.10e-50 | 27 | 322 | 9 | 317 | GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.02e-54 | 34 | 499 | 40 | 496 | Endoglucanase B OS=Paenibacillus lautus OX=1401 GN=celB PE=3 SV=1 |
|
| 1.28e-43 | 28 | 341 | 54 | 381 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
|
| 4.55e-42 | 28 | 321 | 40 | 339 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
|
| 6.10e-42 | 19 | 321 | 30 | 341 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
|
| 1.55e-41 | 17 | 322 | 315 | 602 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
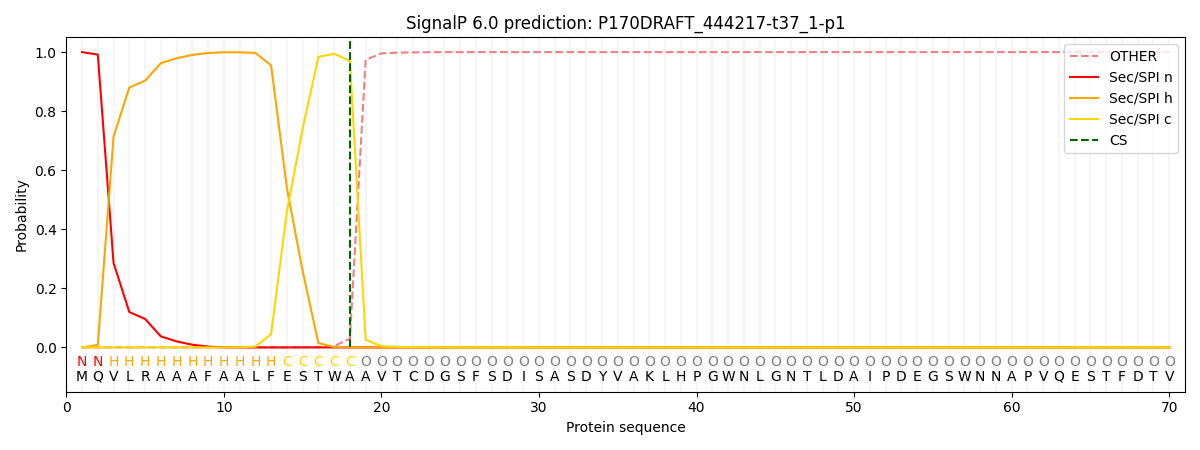
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000415 | 0.999542 | CS pos: 18-19. Pr: 0.9702 |
