You are browsing environment: FUNGIDB
CAZyme Information: P168DRAFT_294406-t37_1-p1
You are here: Home > Sequence: P168DRAFT_294406-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus campestris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus campestris | |||||||||||
| CAZyme ID | P168DRAFT_294406-t37_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | FAD/NAD(P)-binding domain-containing protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 393 | 1022 | 3.6e-93 | 0.9947183098591549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 1.04e-62 | 388 | 1025 | 2 | 538 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 3.87e-62 | 408 | 1020 | 20 | 531 | choline dehydrogenase; Validated |
| 398739 | GMC_oxred_C | 2.68e-32 | 869 | 1016 | 2 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 366272 | GMC_oxred_N | 2.32e-17 | 494 | 769 | 19 | 218 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 399033 | CFEM | 4.94e-17 | 25 | 87 | 3 | 66 | CFEM domain. This fungal specific cysteine rich domain is found in some proteins with proposed roles in fungal pathogenesis. The structure of the CFEM domain containing protein 'Surface antigen protein 2' from Candida albicans has been solved. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 392 | 1024 | 47 | 677 | |
| 0.0 | 392 | 1024 | 47 | 677 | |
| 0.0 | 392 | 1024 | 28 | 658 | |
| 0.0 | 392 | 1024 | 47 | 677 | |
| 0.0 | 405 | 1024 | 49 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.50e-38 | 393 | 1020 | 40 | 597 | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype [Leucoagaricus meleagris] |
|
| 8.15e-37 | 393 | 1020 | 13 | 526 | Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_B Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_C Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_D Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_E Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_F Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_G Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis],3NNE_H Crystal structure of choline oxidase S101A mutant [Arthrobacter globiformis] |
|
| 8.15e-37 | 393 | 1020 | 13 | 526 | Chain A, Choline oxidase [Arthrobacter globiformis],3LJP_B Chain B, Choline oxidase [Arthrobacter globiformis] |
|
| 1.97e-36 | 393 | 1020 | 13 | 526 | Crystal structure of choline oxidase reveals insights into the catalytic mechanism [Arthrobacter globiformis],2JBV_B Crystal structure of choline oxidase reveals insights into the catalytic mechanism [Arthrobacter globiformis],4MJW_A Crystal Structure of Choline Oxidase in Complex with the Reaction Product Glycine Betaine [Arthrobacter globiformis],4MJW_B Crystal Structure of Choline Oxidase in Complex with the Reaction Product Glycine Betaine [Arthrobacter globiformis] |
|
| 8.30e-35 | 393 | 1023 | 1 | 564 | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid [Pleurotus eryngii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.73e-44 | 385 | 1024 | 27 | 611 | Oxidoreductase cicC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cicC PE=1 SV=1 |
|
| 7.34e-42 | 392 | 1023 | 3 | 533 | Oxygen-dependent choline dehydrogenase OS=Photobacterium profundum (strain SS9) OX=298386 GN=betA PE=3 SV=1 |
|
| 1.78e-41 | 381 | 1020 | 32 | 607 | Dehydrogenase pkfF OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pkfF PE=2 SV=1 |
|
| 1.26e-40 | 394 | 1021 | 5 | 531 | Oxygen-dependent choline dehydrogenase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=betA PE=3 SV=1 |
|
| 2.14e-40 | 393 | 1020 | 45 | 576 | Choline dehydrogenase, mitochondrial OS=Rattus norvegicus OX=10116 GN=Chdh PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
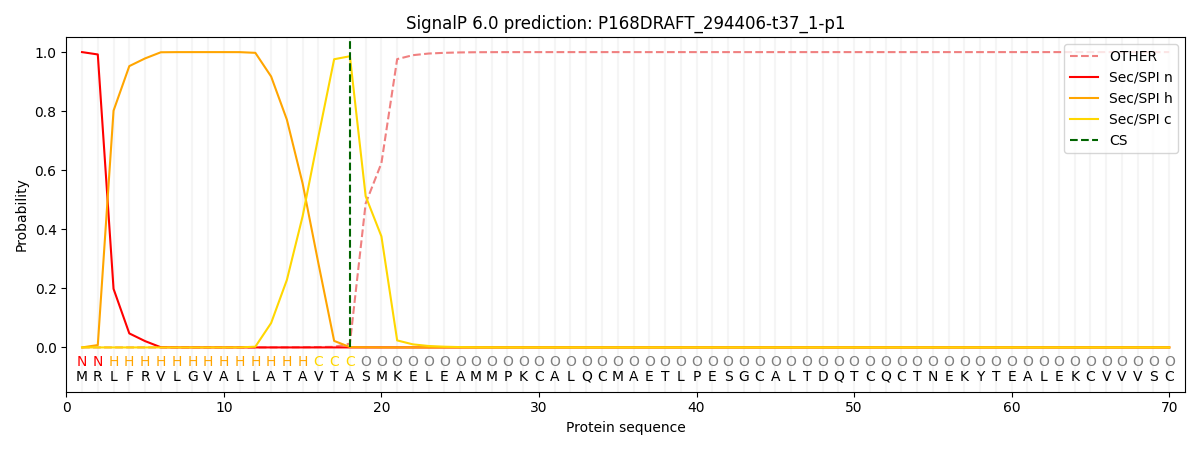
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000286 | 0.999699 | CS pos: 18-19. Pr: 0.9859 |
TMHMM Annotations download full data without filtering help

| Start | End |
|---|---|
| 96 | 118 |
| 125 | 147 |
| 180 | 202 |
| 209 | 231 |
| 255 | 277 |
| 290 | 308 |
