You are browsing environment: FUNGIDB
CAZyme Information: P168DRAFT_280719-t37_1-p1
You are here: Home > Sequence: P168DRAFT_280719-t37_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus campestris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus campestris | |||||||||||
| CAZyme ID | P168DRAFT_280719-t37_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | pectin lyase-like protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 72 | 247 | 3.6e-54 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 239477 | PAP2_dolichyldiphosphatase | 1.05e-39 | 410 | 565 | 3 | 159 | PAP2_like proteins, dolichyldiphosphatase subfamily. Dolichyldiphosphatase is a membrane-associated protein located in the endoplasmic reticulum and hydrolyzes dolichyl pyrophosphate, as well as dolichylmonophosphate at a low rate. The enzyme is necessary for maintaining proper levels of dolichol-linked oligosaccharides and protein N-glycosylation, and might play a role in re-utilization of the glycosyl carrier lipid for additional rounds of lipid intermediate biosynthesis after its release during protein N-glycosylation reactions. |
| 238813 | PAP2_like | 2.98e-10 | 447 | 565 | 1 | 122 | PAP2_like proteins, a super-family of histidine phosphatases and vanadium haloperoxidases, includes type 2 phosphatidic acid phosphatase or lipid phosphate phosphatase (LPP), Glucose-6-phosphatase, Phosphatidylglycerophosphatase B and bacterial acid phosphatase, vanadium chloroperoxidases, vanadium bromoperoxidases, and several other mostly uncharacterized subfamilies. Several members of this superfamily have been predicted to be transmembrane proteins. |
| 226384 | PelB | 4.47e-08 | 8 | 331 | 9 | 344 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 408654 | TSP3_bac | 2.87e-06 | 373 | 393 | 1 | 21 | Bacterial TSP3 repeat. This entry contains a novel bacterial thrombospondin type 3 repeat which differs from the typical consensus by containing a glutamate in place of one of the calcium binding aspartate residues. |
| 214471 | acidPPc | 5.91e-06 | 451 | 565 | 1 | 116 | Acid phosphatase homologues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.32e-230 | 1 | 404 | 1 | 404 | |
| 9.86e-227 | 9 | 404 | 8 | 403 | |
| 1.98e-226 | 9 | 404 | 8 | 403 | |
| 1.98e-226 | 9 | 404 | 8 | 403 | |
| 1.14e-225 | 9 | 404 | 8 | 403 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.17e-233 | 1 | 404 | 1 | 404 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
|
| 8.81e-233 | 1 | 404 | 1 | 404 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
|
| 1.77e-232 | 1 | 404 | 1 | 404 | Probable pectate lyase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyC PE=3 SV=1 |
|
| 6.52e-230 | 4 | 404 | 2 | 403 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
|
| 3.52e-227 | 9 | 404 | 8 | 403 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
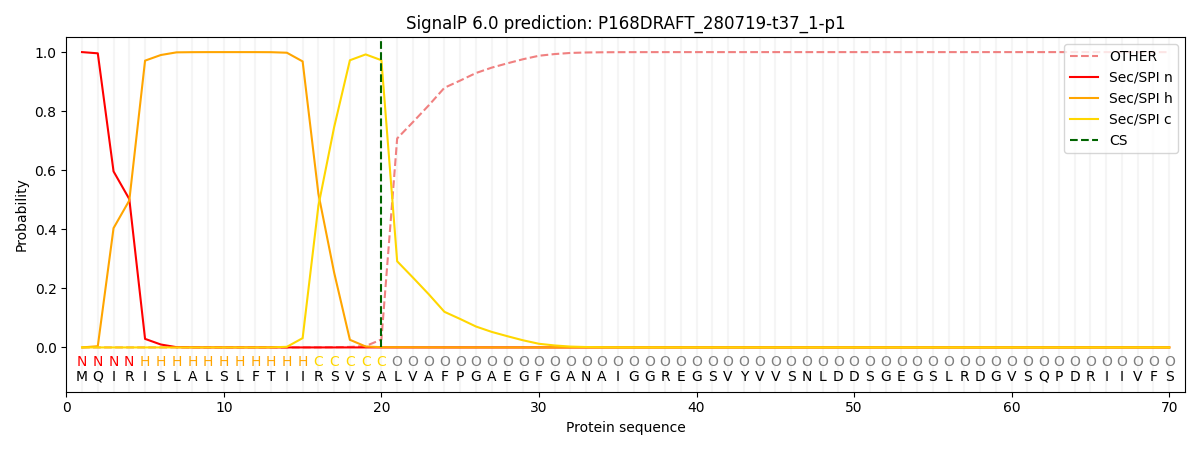
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000616 | 0.999378 | CS pos: 20-21. Pr: 0.9725 |
TMHMM Annotations download full data without filtering help
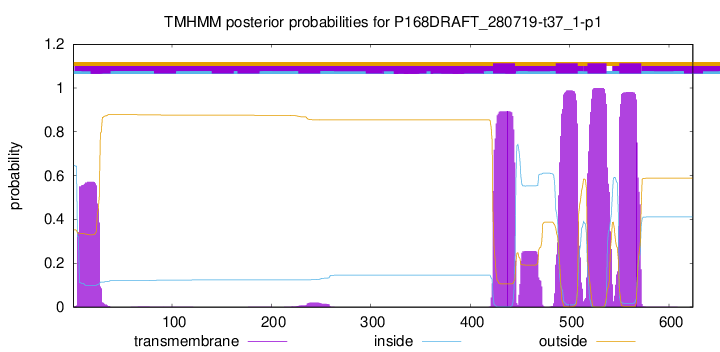
| Start | End |
|---|---|
| 423 | 445 |
| 486 | 508 |
| 518 | 537 |
| 550 | 572 |
