You are browsing environment: FUNGIDB
CAZyme Information: OTA39639.1
You are here: Home > Sequence: OTA39639.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA39639.1 | |||||||||||
| CAZy Family | GT58 | |||||||||||
| CAZyme Description | Glyco_hydro_cc domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TUH8] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 83721; End:84731 Strand: + | |||||||||||
Full Sequence Download help
| MRSTTLLSLA AASLPLALAQ DDSLTSSKRG LCYVDPEENT QDTDVWDASD SDLTWYYNYE | 60 |
| ASPTDGIDES KLQFVPMIWG LPDSTSSMAF YNTVTGLMEQ GYNITYVLGF NEPDGCVSGG | 120 |
| SCIAAEDAAA AWIREIEPLK DQGVLLGAPA VTGSPMGFTW LENFYNACNG NCTTHFIPVH | 180 |
| WYGNFEGLAS HMGQVNGTYP NMTMWVTEYA CADCELEEAQ QFYNMSADYF DRLDYVTHYS | 240 |
| YFGAFRSDVS NVGKNAAMLT QKGELTDIGA WYLGQQATGN KPKGGAAQVA KFAGWAVVVA | 300 |
| SVAFWTVL | 308 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH128 | 47 | 272 | 7.4e-68 | 0.9821428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 371727 | Glyco_hydro_cc | 9.84e-72 | 39 | 272 | 4 | 235 | Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UJO21606.1|GH128 | 1.34e-118 | 13 | 307 | 11 | 304 |
| SMY25379.1|GH128 | 1.05e-116 | 25 | 307 | 29 | 309 |
| SMR54023.1|GH128 | 1.05e-116 | 25 | 307 | 29 | 309 |
| SMQ51738.1|GH128 | 1.05e-116 | 25 | 307 | 29 | 309 |
| SMR56196.1|GH128 | 1.05e-116 | 25 | 307 | 29 | 309 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UBC_A | 6.14e-14 | 53 | 273 | 219 | 437 | Chain A, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q09788|ASL1_SCHPO | 1.99e-20 | 23 | 273 | 291 | 529 | Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
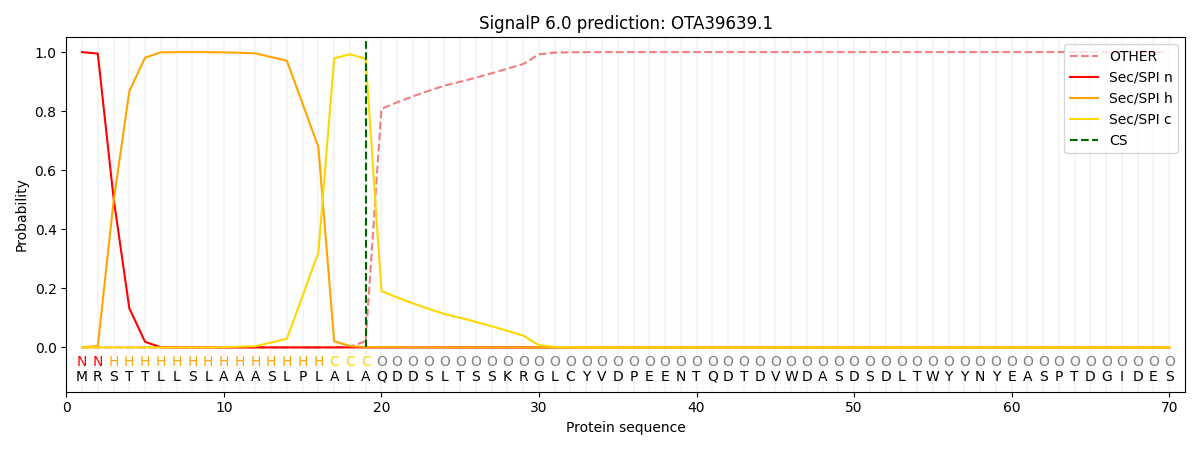
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000274 | 0.999687 | CS pos: 19-20. Pr: 0.9774 |
