You are browsing environment: FUNGIDB
CAZyme Information: OTA39240.1
You are here: Home > Sequence: OTA39240.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA39240.1 | |||||||||||
| CAZy Family | GT4 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 50 | 682 | 1.1e-96 | 0.9929577464788732 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235000 | PRK02106 | 3.80e-54 | 65 | 681 | 20 | 531 | choline dehydrogenase; Validated |
| 225186 | BetA | 7.74e-50 | 65 | 681 | 22 | 533 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 398739 | GMC_oxred_C | 4.43e-28 | 536 | 677 | 5 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 215420 | PLN02785 | 1.24e-17 | 317 | 663 | 230 | 558 | Protein HOTHEAD |
| 366272 | GMC_oxred_N | 2.30e-11 | 151 | 425 | 19 | 218 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.58e-302 | 24 | 685 | 19 | 677 | |
| 5.58e-302 | 24 | 685 | 19 | 677 | |
| 5.58e-302 | 24 | 685 | 19 | 677 | |
| 5.63e-302 | 46 | 685 | 25 | 658 | |
| 7.63e-302 | 49 | 685 | 45 | 673 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.43e-40 | 50 | 681 | 40 | 597 | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype [Leucoagaricus meleagris] |
|
| 4.21e-35 | 50 | 681 | 7 | 560 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis] |
|
| 5.67e-35 | 50 | 681 | 7 | 560 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis] |
|
| 7.63e-35 | 50 | 681 | 7 | 560 | Chain A, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis],6ZH7_B Chain B, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis] |
|
| 7.63e-35 | 50 | 681 | 7 | 560 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRV_AAA Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRX_AAA Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRZ_AAA Chain AAA, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.89e-41 | 48 | 679 | 4 | 533 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas syringae pv. tomato (strain ATCC BAA-871 / DC3000) OX=223283 GN=betA PE=3 SV=1 |
|
| 4.88e-41 | 51 | 682 | 36 | 608 | Oxidoreductase cicC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cicC PE=1 SV=1 |
|
| 1.22e-40 | 40 | 685 | 30 | 611 | Dehydrogenase pkfF OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pkfF PE=2 SV=1 |
|
| 6.05e-40 | 50 | 679 | 4 | 531 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=betA PE=3 SV=1 |
|
| 1.13e-39 | 48 | 679 | 4 | 533 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=betA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
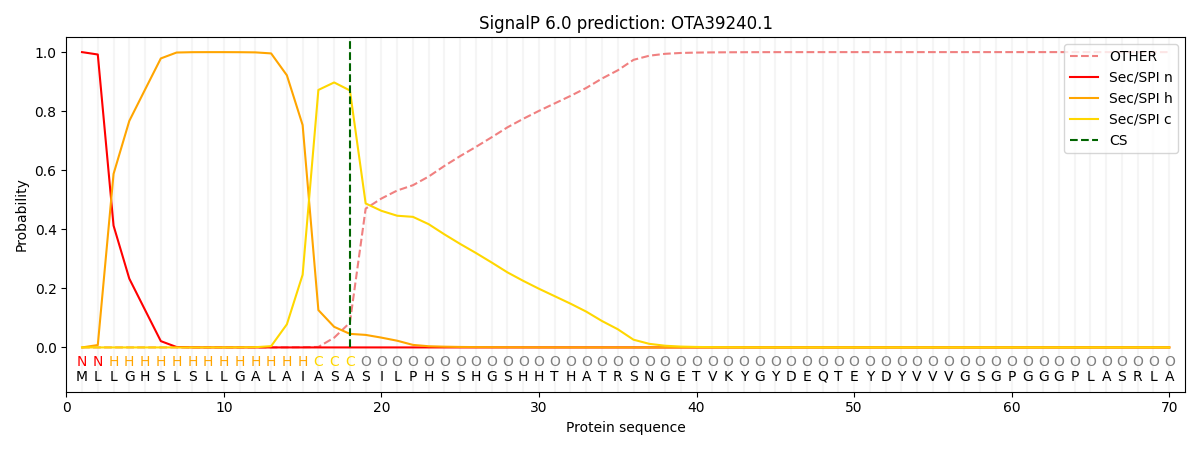
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001087 | 0.998867 | CS pos: 18-19. Pr: 0.8702 |
