You are browsing environment: FUNGIDB
CAZyme Information: OTA37910.1
You are here: Home > Sequence: OTA37910.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
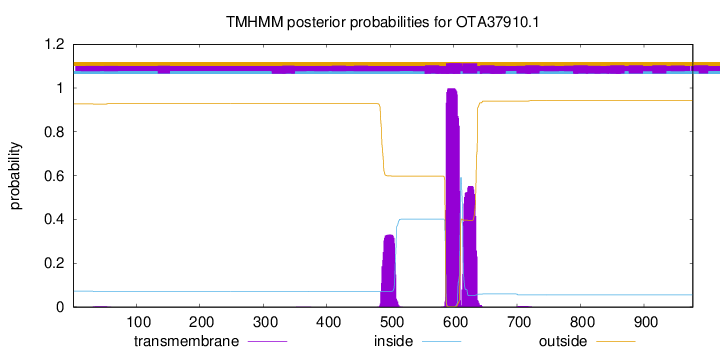
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA37910.1 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | Brix domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TPG4] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 621 | 929 | 1.1e-80 | 0.983739837398374 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227437 | RPF2 | 2.28e-100 | 4 | 301 | 2 | 292 | Uncharacterized conserved protein [Function unknown]. |
| 368535 | Glyco_transf_34 | 1.67e-75 | 620 | 927 | 3 | 238 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 398230 | Brix | 1.28e-31 | 38 | 241 | 2 | 137 | Brix domain. |
| 214879 | Brix | 1.36e-27 | 33 | 237 | 1 | 180 | The Brix domain is found in a number of eukaryotic proteins. Members include SSF proteins from yeast and humans, Arabidopsis thaliana Peter Pan-like protein and several hypothetical proteins. |
| 215616 | PLN03181 | 7.25e-04 | 683 | 725 | 183 | 225 | glycosyltransferase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.06e-237 | 436 | 957 | 1 | 487 | |
| 1.48e-201 | 436 | 957 | 1 | 493 | |
| 1.48e-201 | 436 | 957 | 1 | 493 | |
| 1.48e-201 | 436 | 957 | 1 | 493 | |
| 1.44e-194 | 436 | 954 | 1 | 468 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.50e-81 | 4 | 308 | 2 | 303 | Cryo-em structure of eukaryotic pre-60S ribosomal subunits [Saccharomyces cerevisiae S288C],6FT6_v Chain v, Ribosome biogenesis protein RPF2 [Saccharomyces cerevisiae S288C],7BT6_v Cryo-EM structure of pre-60S ribosome from Saccharomyces cerevisiae rpl4delta63-87 strain at 3.12 Angstroms resolution(state R1) [Saccharomyces cerevisiae S288C],7BTB_v Cryo-EM structure of pre-60S ribosome from Saccharomyces cerevisiae rpl4delta63-87 strain at 3.22 Angstroms resolution(state R2) [Saccharomyces cerevisiae S288C],7OH3_v Chain v, Ribosome biogenesis protein RPF2 [Saccharomyces cerevisiae S288C],7OHQ_v Chain v, Ribosome biogenesis protein RPF2 [Saccharomyces cerevisiae S288C],7OHT_v Chain v, Ribosome biogenesis protein RPF2 [Saccharomyces cerevisiae S288C] |
|
| 1.18e-78 | 4 | 300 | 23 | 320 | Structure of Rpf2-Rrs1 complex involved in ribosome biogenesis [Aspergillus nidulans FGSC A4],4XD9_C Structure of Rpf2-Rrs1 complex involved in ribosome biogenesis [Aspergillus nidulans FGSC A4] |
|
| 1.89e-71 | 20 | 293 | 1 | 271 | Crystal structure of the Rrs1 and Rpf2 complex [Saccharomyces cerevisiae S288C],5WXL_C Crystal structure of the Rrs1 and Rpf2 complex [Saccharomyces cerevisiae S288C] |
|
| 1.67e-52 | 25 | 257 | 1 | 230 | Crystal Structure of the Rpf2-Rrs1 complex [Saccharomyces cerevisiae] |
|
| 2.27e-40 | 25 | 256 | 1 | 240 | The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis [Aspergillus nidulans FGSC A4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.37e-90 | 4 | 307 | 2 | 299 | Ribosome production factor 2 homolog OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC926.08c PE=1 SV=1 |
|
| 1.04e-86 | 621 | 943 | 121 | 384 | Probable alpha-1,6-mannosyltransferase MNN10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN10 PE=1 SV=1 |
|
| 2.31e-80 | 4 | 308 | 2 | 303 | Ribosome biogenesis protein RPF2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=RPF2 PE=1 SV=1 |
|
| 5.30e-70 | 620 | 949 | 73 | 343 | Uncharacterized alpha-1,2-galactosyltransferase C637.06 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC637.06 PE=2 SV=2 |
|
| 3.18e-56 | 4 | 308 | 4 | 294 | Ribosome production factor 2 homolog OS=Drosophila melanogaster OX=7227 GN=Non3 PE=2 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000068 | 0.000000 |