You are browsing environment: FUNGIDB
CAZyme Information: OTA37326.1
You are here: Home > Sequence: OTA37326.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA37326.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | Chitin synthase [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TMT0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.16:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224136 | BcsA | 2.85e-15 | 34 | 396 | 9 | 397 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 276809 | TPR | 3.73e-14 | 893 | 973 | 1 | 81 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 133045 | CESA_like | 1.10e-11 | 85 | 269 | 1 | 152 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| 276809 | TPR | 6.67e-11 | 867 | 955 | 10 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 223533 | TPR | 6.69e-07 | 769 | 977 | 83 | 282 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.69e-257 | 10 | 487 | 62 | 605 | |
| 1.57e-256 | 1 | 495 | 74 | 629 | |
| 9.16e-256 | 1 | 495 | 74 | 629 | |
| 1.69e-255 | 1 | 495 | 74 | 629 | |
| 6.95e-255 | 1 | 495 | 74 | 629 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.90e-197 | 8 | 485 | 29 | 566 | Chitin synthase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=chsD PE=3 SV=1 |
|
| 6.36e-113 | 516 | 1230 | 136 | 817 | TPR repeat-containing protein C19B12.01 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC19B12.01 PE=4 SV=2 |
|
| 1.24e-73 | 514 | 1224 | 193 | 899 | Essential for maintenance of the cell wall protein 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=EMW1 PE=1 SV=1 |
|
| 2.68e-47 | 501 | 1226 | 174 | 839 | Tetratricopeptide repeat protein 27 homolog OS=Dictyostelium discoideum OX=44689 GN=ttc27 PE=3 SV=1 |
|
| 3.86e-42 | 508 | 943 | 179 | 581 | Tetratricopeptide repeat protein 27 OS=Bos taurus OX=9913 GN=TTC27 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999998 | 0.000007 |
TMHMM Annotations download full data without filtering help
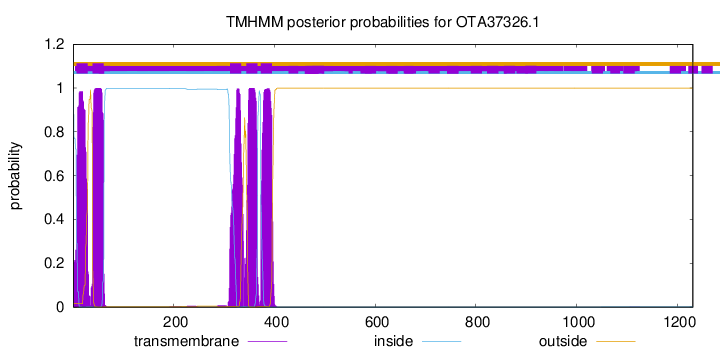
| Start | End |
|---|---|
| 7 | 29 |
| 39 | 61 |
| 312 | 334 |
| 344 | 366 |
| 373 | 395 |
