You are browsing environment: FUNGIDB
CAZyme Information: OTA36925.1
You are here: Home > Sequence: OTA36925.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA36925.1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | Amb_all domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TLQ7] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 4.2.2.10:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 111 | 296 | 8.1e-86 | 0.983957219251337 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 3.32e-42 | 111 | 299 | 2 | 190 | Amb_all domain. |
| 226384 | PelB | 6.92e-17 | 66 | 293 | 28 | 271 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 8.16e-14 | 123 | 293 | 32 | 209 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 369468 | MFS_1 | 6.56e-09 | 408 | 523 | 1 | 117 | Major Facilitator Superfamily. |
| 349949 | MFS | 4.76e-08 | 408 | 523 | 1 | 116 | Major Facilitator Superfamily. The Major Facilitator Superfamily (MFS) is a large and diverse group of secondary transporters that includes uniporters, symporters, and antiporters. MFS proteins facilitate the transport across cytoplasmic or internal membranes of a variety of substrates including ions, sugar phosphates, drugs, neurotransmitters, nucleosides, amino acids, and peptides. They do so using the electrochemical potential of the transported substrates. Uniporters transport a single substrate, while symporters and antiporters transport two substrates in the same or in opposite directions, respectively, across membranes. MFS proteins are typically 400 to 600 amino acids in length, and the majority contain 12 transmembrane alpha helices (TMs) connected by hydrophilic loops. The N- and C-terminal halves of these proteins display weak similarity and may be the result of a gene duplication/fusion event. Based on kinetic studies and the structures of a few bacterial superfamily members, GlpT (glycerol-3-phosphate transporter), LacY (lactose permease), and EmrD (multidrug transporter), MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. Bacterial members function primarily for nutrient uptake, and as drug-efflux pumps to confer antibiotic resistance. Some MFS proteins have medical significance in humans such as the glucose transporter Glut4, which is impaired in type II diabetes, and glucose-6-phosphate transporter (G6PT), which causes glycogen storage disease when mutated. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.36e-133 | 22 | 392 | 22 | 383 | |
| 4.31e-132 | 19 | 354 | 19 | 353 | |
| 4.31e-132 | 19 | 354 | 19 | 353 | |
| 6.17e-123 | 19 | 354 | 19 | 351 | |
| 1.40e-121 | 15 | 343 | 16 | 340 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.81e-99 | 21 | 356 | 3 | 332 | Pectin Lyase A [Aspergillus niger] |
|
| 1.04e-96 | 21 | 356 | 3 | 332 | Pectin Lyase A [Aspergillus niger],1IDJ_B Pectin Lyase A [Aspergillus niger] |
|
| 6.78e-90 | 19 | 357 | 1 | 334 | Pectin Lyase B [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.58e-121 | 21 | 346 | 23 | 343 | Probable pectin lyase F-2 OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pelF-2 PE=3 SV=1 |
|
| 1.83e-120 | 18 | 365 | 19 | 358 | Probable pectin lyase F OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pelF PE=3 SV=1 |
|
| 1.30e-118 | 21 | 343 | 23 | 342 | Probable pectin lyase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pelF PE=3 SV=2 |
|
| 1.30e-118 | 21 | 343 | 23 | 342 | Probable pectin lyase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=pelF PE=3 SV=1 |
|
| 2.36e-117 | 21 | 342 | 24 | 342 | Probable pectin lyase F OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pelF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
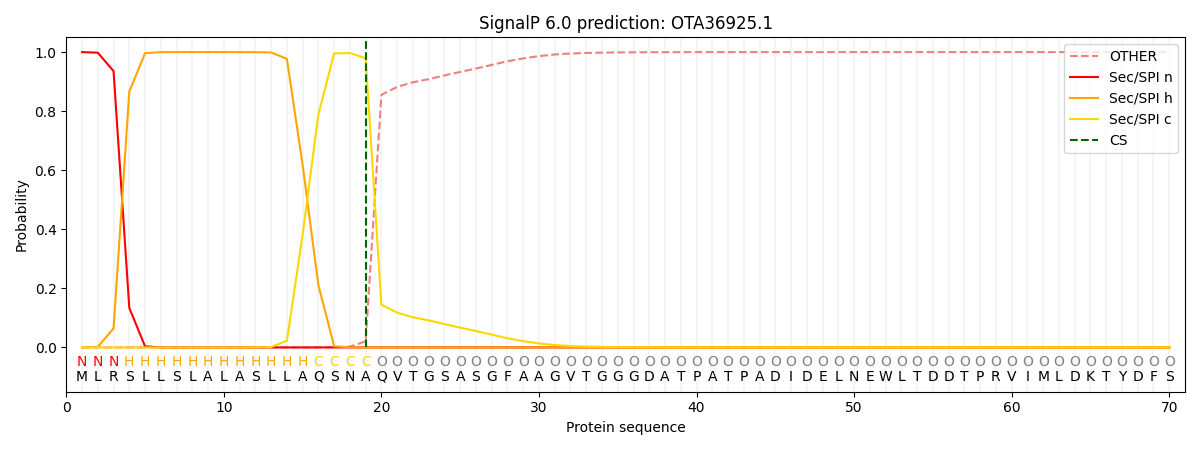
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000279 | 0.999701 | CS pos: 19-20. Pr: 0.9792 |
TMHMM Annotations download full data without filtering help
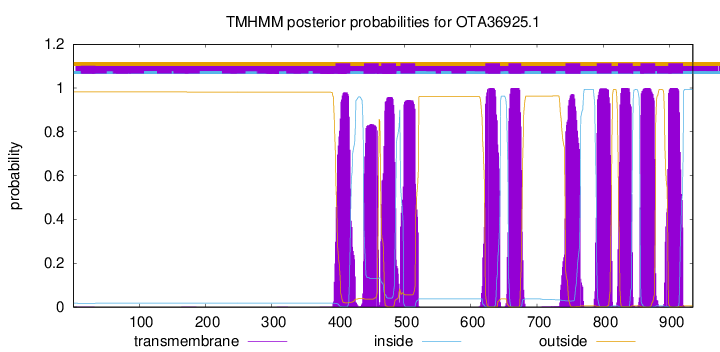
| Start | End |
|---|---|
| 396 | 418 |
| 439 | 461 |
| 465 | 487 |
| 494 | 516 |
| 622 | 644 |
| 657 | 676 |
| 743 | 765 |
| 790 | 812 |
| 822 | 844 |
| 856 | 878 |
| 898 | 920 |
