You are browsing environment: FUNGIDB
CAZyme Information: OTA36020.1
You are here: Home > Sequence: OTA36020.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA36020.1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | Beta-N-acetylhexosaminidase [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TJ47] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | - | 3.2.1.52:9 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 408 | 754 | 1.8e-87 | 0.9762611275964391 |
| GH16 | 76 | 230 | 4e-71 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119332 | GH20_HexA_HexB-like | 8.84e-146 | 414 | 783 | 1 | 348 | Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| 395590 | Glyco_hydro_20 | 2.28e-109 | 414 | 754 | 1 | 345 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| 185692 | GH16_fungal_CRH1_transglycosylase | 6.51e-102 | 55 | 252 | 1 | 203 | glycosylphosphatidylinositol-glucanosyltransferase. Group of fungal GH16 members related to Saccharomyces cerevisiae Crh1p. Chr1p and Crh2p are transglycosylases that are required for the linkage of chitin to beta(1-3)glucose branches of beta(1-6)glucan, an important step in the assembly of new cell wall. Both have been shown to be glycosylphosphatidylinositol (GPI)-anchored. A third homologous protein, Crr1p, functions in the formation of the spore wall. They belongs to the family 16 of glycosyl hydrolases that includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| 119333 | GH20_chitobiase-like | 1.78e-68 | 414 | 756 | 1 | 346 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| 119331 | GH20_hexosaminidase | 7.67e-58 | 416 | 753 | 1 | 303 | Beta-N-acetylhexosaminidases of glycosyl hydrolase family 20 (GH20) catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. These enzymes are broadly distributed in microorganisms, plants and animals, and play roles in various key physiological and pathological processes. These processes include cell structural integrity, energy storage, cellular signaling, fertilization, pathogen defense, viral penetration, the development of carcinomas, inflammatory events and lysosomal storage disorders. The GH20 enzymes include the eukaryotic beta-N-acetylhexosaminidases A and B, the bacterial chitobiases, dispersin B, and lacto-N-biosidase. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by the solvent or the enzyme, but by the substrate itself. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.96e-251 | 330 | 803 | 92 | 562 | |
| 3.02e-249 | 324 | 802 | 96 | 572 | |
| 6.07e-249 | 324 | 802 | 96 | 572 | |
| 6.07e-249 | 324 | 802 | 96 | 572 | |
| 6.07e-249 | 324 | 802 | 96 | 572 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.71e-78 | 32 | 270 | 1 | 243 | Apo Crh5 transglycosylase [Aspergillus fumigatus Af293],6IBU_B Apo Crh5 transglycosylase [Aspergillus fumigatus Af293],6IBW_A Crh5 transglycosylase in complex with NAG [Aspergillus fumigatus Af293],6IBW_B Crh5 transglycosylase in complex with NAG [Aspergillus fumigatus Af293] |
|
| 3.76e-70 | 355 | 803 | 23 | 491 | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae [Aspergillus oryzae],5OAR_D Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae [Aspergillus oryzae] |
|
| 4.10e-62 | 355 | 801 | 125 | 570 | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis [Ostrinia furnacalis],3NSN_A Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin [Ostrinia furnacalis],3OZO_A Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT [Ostrinia furnacalis],3OZP_A Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc [Ostrinia furnacalis] |
|
| 4.10e-62 | 355 | 801 | 125 | 570 | Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q1 [Ostrinia furnacalis],3WMC_A Crystal structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with naphthalimide derivative Q2 [Ostrinia furnacalis] |
|
| 4.36e-62 | 355 | 801 | 128 | 573 | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 V327G complexed with PUGNAc [Ostrinia furnacalis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.51e-131 | 356 | 803 | 156 | 605 | Beta-hexosaminidase ARB_07893 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07893 PE=1 SV=1 |
|
| 2.71e-80 | 14 | 277 | 1 | 264 | Probable extracellular glycosidase ARB_05253 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_05253 PE=1 SV=2 |
|
| 1.03e-79 | 332 | 801 | 91 | 570 | Beta-hexosaminidase 2 OS=Arabidopsis thaliana OX=3702 GN=HEXO2 PE=1 SV=1 |
|
| 5.72e-77 | 29 | 273 | 19 | 267 | Probable glycosidase crf1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=crf1 PE=1 SV=2 |
|
| 1.55e-75 | 16 | 278 | 6 | 269 | Extracellular glycosidase CRH11 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CRH11 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
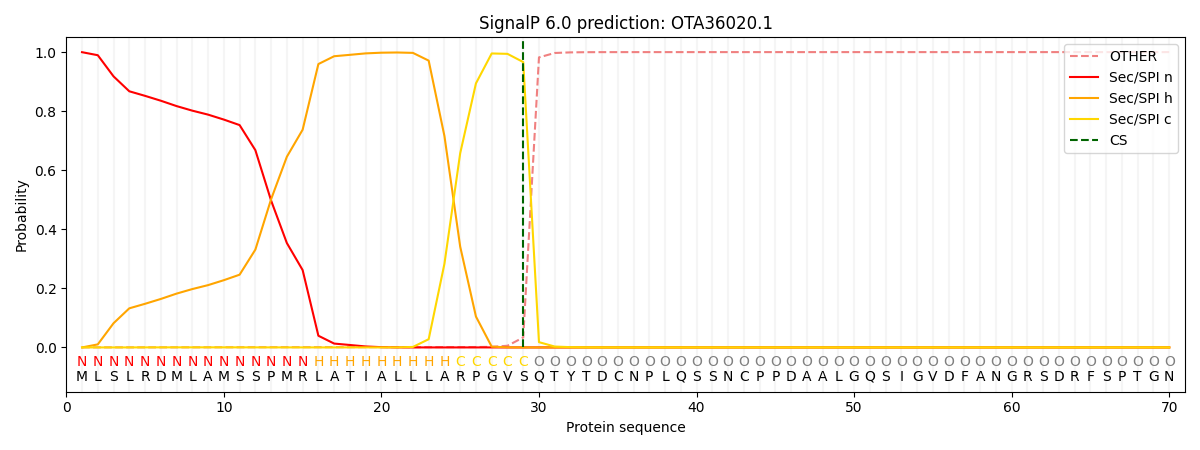
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000788 | 0.999198 | CS pos: 29-30. Pr: 0.9663 |
