You are browsing environment: FUNGIDB
CAZyme Information: OTA35373.1
You are here: Home > Sequence: OTA35373.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA35373.1 | |||||||||||
| CAZy Family | GH62 | |||||||||||
| CAZyme Description | 1,3-beta-glucanosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TH77] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:26 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH72 | 44 | 327 | 9.1e-120 | 0.9358974358974359 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 239238 | FCB2_FMN | 0.0 | 476 | 821 | 2 | 344 | Flavocytochrome b2 (FCB2) FMN-binding domain. FCB2 (AKA L-lactate:cytochrome c oxidoreductase) is a respiratory enzyme located in the intermembrane space of fungal mitochondria which catalyzes the oxidation of L-lactate to pyruvate. FCB2 also participates in a short electron-transport chain involving cytochrome c and cytochrome oxidase which ultimately directs the reducing equivalents gained from L-lactate oxidation to oxygen, yielding one molecule of ATP for every L-lactate molecule consumed. FCB2 is composed of 2 domains: a C-terminal flavin-binding domain, which includes the active site for lacate oxidation, and an N-terminal b2-cytochrome domain, required for efficient cytochrome c reduction. FCB2 is a homotetramer and contains two noncovalently bound cofactors, FMN and heme per subunit. |
| 397351 | Glyco_hydro_72 | 1.64e-158 | 45 | 345 | 9 | 315 | Glucanosyltransferase. This is a family of glycosylphosphatidylinositol-anchored beta(1-3)glucanosyltransferases. The active site residues in the Aspergillus fumigatus example are the two glutamate residues at 160 and 261. |
| 395850 | FMN_dh | 1.76e-152 | 481 | 825 | 1 | 350 | FMN-dependent dehydrogenase. |
| 239203 | alpha_hydroxyacid_oxid_FMN | 7.18e-130 | 476 | 820 | 2 | 299 | Family of homologous FMN-dependent alpha-hydroxyacid oxidizing enzymes. This family occurs in both prokaryotes and eukaryotes. Members of this family include flavocytochrome b2 (FCB2), glycolate oxidase (GOX), lactate monooxygenase (LMO), mandelate dehydrogenase (MDH), and long chain hydroxyacid oxidase (LCHAO). In green plants, glycolate oxidase is one of the key enzymes in photorespiration where it oxidizes glycolate to glyoxylate. LMO catalyzes the oxidation of L-lactate to acetate and carbon dioxide. MDH oxidizes (S)-mandelate to phenylglyoxalate. It is an enzyme in the mandelate pathway that occurs in several strains of Pseudomonas which converts (R)-mandelate to benzoate. |
| 224223 | LldD | 2.93e-105 | 474 | 823 | 1 | 348 | FMN-dependent dehydrogenase, includes L-lactate dehydrogenase and type II isopentenyl diphosphate isomerase [Energy production and conversion, Lipid transport and metabolism, General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.64e-173 | 24 | 384 | 24 | 385 | |
| 2.31e-151 | 14 | 370 | 10 | 358 | |
| 2.31e-151 | 14 | 370 | 10 | 358 | |
| 1.60e-149 | 41 | 383 | 33 | 374 | |
| 1.24e-148 | 41 | 370 | 33 | 361 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.79e-152 | 393 | 835 | 33 | 485 | The 2.6 Angstroms Refined Structure Of The Escherichia Coli Recombinant Saccharomyces Cerevisiae Flavocytochrome B2-Sulphite Complex [Saccharomyces cerevisiae],1LTD_B The 2.6 Angstroms Refined Structure Of The Escherichia Coli Recombinant Saccharomyces Cerevisiae Flavocytochrome B2-Sulphite Complex [Saccharomyces cerevisiae] |
|
| 1.04e-151 | 393 | 835 | 38 | 490 | Molecular Structure Of Flavocytochrome B2 At 2.4 Angstroms Resolution [Saccharomyces cerevisiae],1FCB_B Molecular Structure Of Flavocytochrome B2 At 2.4 Angstroms Resolution [Saccharomyces cerevisiae],1KBI_A Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme [Saccharomyces cerevisiae],1KBI_B Crystallographic Study of the Recombinant Flavin-binding Domain of Baker's Yeast Flavocytochrome b2: Comparison with the Intact Wild-type Enzyme [Saccharomyces cerevisiae] |
|
| 4.07e-151 | 393 | 835 | 38 | 490 | X-RAY STRUCTURE OF TWO COMPLEXES OF THE Y143F FLAVOCYTOCHROME B2 MUTANT CRYSTALLIZED IN THE PRESENCE OF LACTATE OR PHENYL-LACTATE [Saccharomyces cerevisiae],1LCO_B X-RAY STRUCTURE OF TWO COMPLEXES OF THE Y143F FLAVOCYTOCHROME B2 MUTANT CRYSTALLIZED IN THE PRESENCE OF LACTATE OR PHENYL-LACTATE [Saccharomyces cerevisiae],1LDC_A X-RAY STRUCTURE OF TWO COMPLEXES OF THE Y143F FLAVOCYTOCHROME B2 MUTANT CRYSTALLIZED IN THE PRESENCE OF LACTATE OR PHENYL-LACTATE [Saccharomyces cerevisiae],1LDC_B X-RAY STRUCTURE OF TWO COMPLEXES OF THE Y143F FLAVOCYTOCHROME B2 MUTANT CRYSTALLIZED IN THE PRESENCE OF LACTATE OR PHENYL-LACTATE [Saccharomyces cerevisiae] |
|
| 5.73e-151 | 393 | 835 | 38 | 490 | Chain A, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae],1SZE_B Chain B, Cytochrome b2, mitochondrial [Saccharomyces cerevisiae] |
|
| 1.59e-150 | 393 | 835 | 38 | 490 | Chain A, Cytochrome b2 [Saccharomyces cerevisiae],2OZ0_B Chain B, Cytochrome b2 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.07e-182 | 396 | 837 | 113 | 551 | L-lactate dehydrogenase (cytochrome) OS=Wickerhamomyces anomalus OX=4927 GN=CYB2 PE=1 SV=2 |
|
| 7.22e-150 | 393 | 835 | 118 | 570 | L-lactate dehydrogenase (cytochrome) OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CYB2 PE=1 SV=1 |
|
| 6.94e-132 | 1 | 384 | 1 | 367 | 1,3-beta-glucanosyltransferase gel1 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=gel1 PE=1 SV=1 |
|
| 1.05e-130 | 1 | 384 | 1 | 367 | 1,3-beta-glucanosyltransferase gel1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=gel1 PE=3 SV=1 |
|
| 3.06e-112 | 40 | 386 | 22 | 371 | 1,3-beta-glucanosyltransferase GAS5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GAS5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
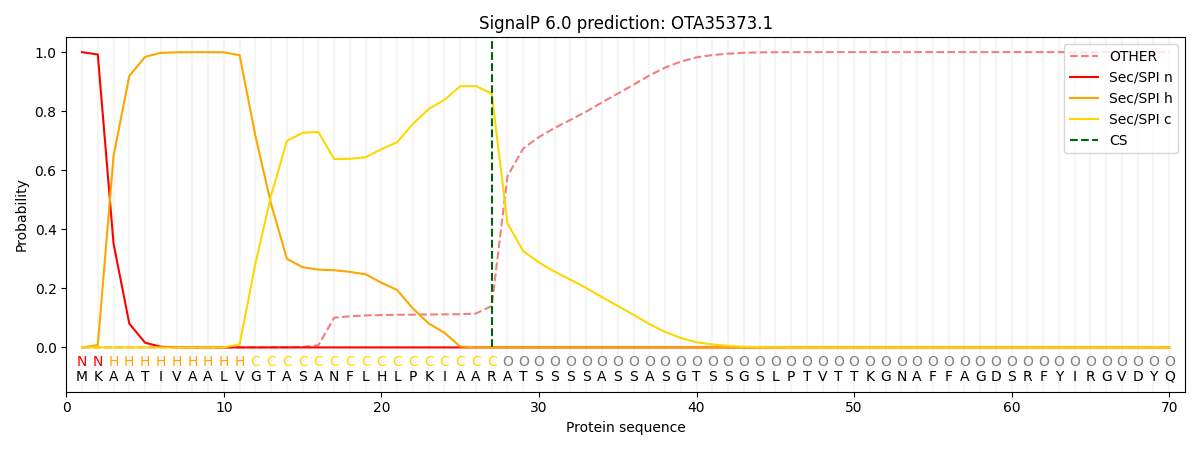
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000869 | 0.999091 | CS pos: 27-28. Pr: 0.8594 |
