You are browsing environment: FUNGIDB
CAZyme Information: OTA34888.1
You are here: Home > Sequence: OTA34888.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA34888.1 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | CAP10 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TFU5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214773 | CAP10 | 4.69e-23 | 162 | 390 | 4 | 250 | Putative lipopolysaccharide-modifying enzyme. |
| 310354 | Glyco_transf_90 | 4.18e-20 | 162 | 386 | 81 | 316 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.43e-76 | 50 | 418 | 76 | 448 | |
| 8.17e-76 | 58 | 412 | 89 | 452 | |
| 8.17e-76 | 58 | 412 | 89 | 452 | |
| 1.26e-74 | 58 | 412 | 200 | 563 | |
| 1.54e-71 | 58 | 392 | 70 | 411 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.48e-14 | 162 | 390 | 96 | 339 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
|
| 1.50e-10 | 185 | 389 | 155 | 375 | Crystal structure of Drosophila Poglut1 (Rumi) complexed with its glycoprotein product (glucosylated EGF repeat) and UDP [Drosophila melanogaster],5F85_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) and UDP [Drosophila melanogaster],5F86_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) [Drosophila melanogaster],5F87_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_B Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_C Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_D Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_E Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_F Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.33e-14 | 131 | 390 | 96 | 367 | Protein O-glucosyltransferase 1 OS=Rattus norvegicus OX=10116 GN=Poglut1 PE=3 SV=1 |
|
| 1.28e-13 | 124 | 390 | 90 | 367 | Protein O-glucosyltransferase 1 OS=Mus musculus OX=10090 GN=Poglut1 PE=1 SV=2 |
|
| 4.10e-13 | 162 | 390 | 124 | 367 | Protein O-glucosyltransferase 1 OS=Bos taurus OX=9913 GN=POGLUT1 PE=2 SV=1 |
|
| 4.10e-13 | 162 | 390 | 124 | 367 | Protein O-glucosyltransferase 1 OS=Homo sapiens OX=9606 GN=POGLUT1 PE=1 SV=1 |
|
| 4.31e-12 | 202 | 405 | 177 | 398 | O-glucosyltransferase rumi homolog OS=Anopheles gambiae OX=7165 GN=AGAP004267 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
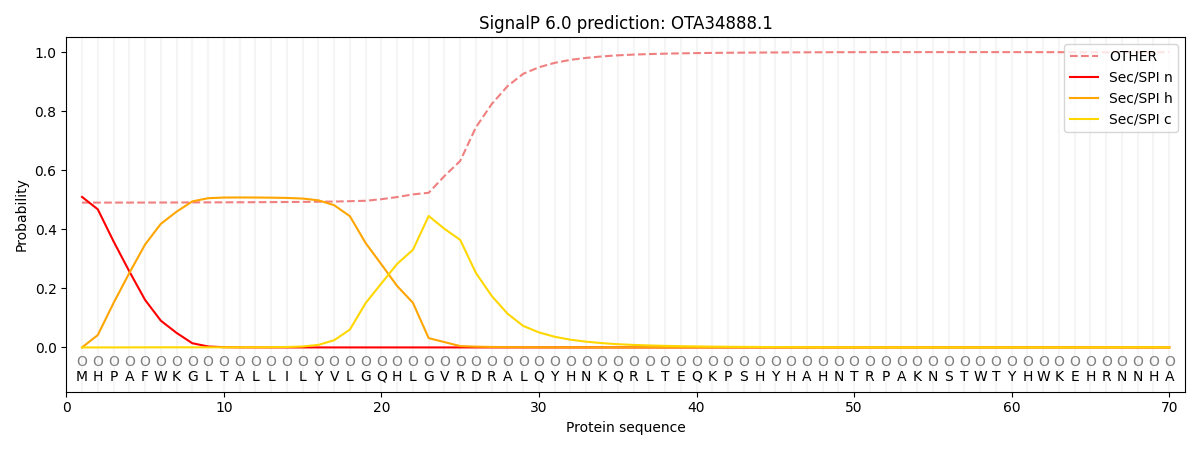
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.506289 | 0.493718 |
