You are browsing environment: FUNGIDB
CAZyme Information: OTA34636.1
You are here: Home > Sequence: OTA34636.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
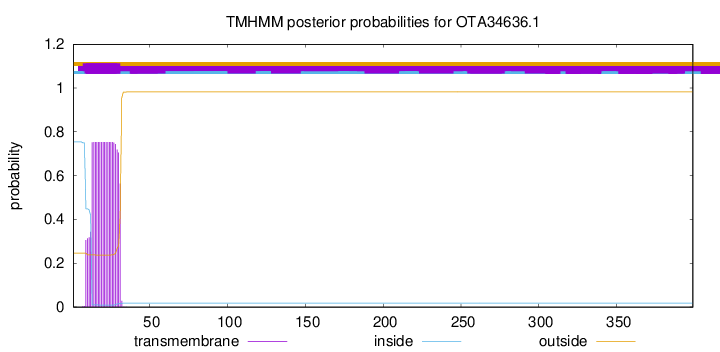
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA34636.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT8 | 4 | 271 | 2.7e-34 | 0.8949416342412452 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133018 | GT8_Glycogenin | 1.61e-67 | 9 | 299 | 1 | 240 | Glycogenin belongs the GT 8 family and initiates the biosynthesis of glycogen. Glycogenin initiates the biosynthesis of glycogen by incorporating glucose residues through a self-glucosylation reaction at a Tyr residue, and then acts as substrate for chain elongation by glycogen synthase and branching enzyme. It contains a conserved DxD motif and an N-terminal beta-alpha-beta Rossmann-like fold that are common to the nucleotide-binding domains of most glycosyltransferases. The DxD motif is essential for coordination of the catalytic divalent cation, most commonly Mn2+. Glycogenin can be classified as a retaining glycosyltransferase, based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. It is placed in glycosyltransferase family 8 which includes lipopolysaccharide glucose and galactose transferases and galactinol synthases. |
| 215090 | PLN00176 | 2.40e-20 | 2 | 271 | 16 | 267 | galactinol synthase |
| 227884 | Gnt1 | 9.41e-16 | 8 | 271 | 70 | 352 | Alpha-N-acetylglucosamine transferase [Cell wall/membrane/envelope biogenesis]. |
| 133064 | GT8_GNT1 | 1.11e-13 | 9 | 124 | 1 | 113 | GNT1 is a fungal enzyme that belongs to the GT 8 family. N-acetylglucosaminyltransferase is a fungal enzyme that catalyzes the addition of N-acetyl-D-glucosamine to mannotetraose side chains by an alpha 1-2 linkage during the synthesis of mannan. The N-acetyl-D-glucosamine moiety in mannan plays a role in the attachment of mannan to asparagine residues in proteins. The mannotetraose and its N-acetyl-D-glucosamine derivative side chains of mannan are the principle immunochemical determinants on the cell surface. N-acetylglucosaminyltransferase is a member of glycosyltransferase family 8, which are, based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed, retaining glycosyltransferases. |
| 132996 | Glyco_transf_8 | 8.13e-06 | 9 | 229 | 1 | 197 | Members of glycosyltransferase family 8 (GT-8) are involved in lipopolysaccharide biosynthesis and glycogen synthesis. Members of this family are involved in lipopolysaccharide biosynthesis and glycogen synthesis. GT-8 comprises enzymes with a number of known activities: lipopolysaccharide galactosyltransferase, lipopolysaccharide glucosyltransferase 1, glycogenin glucosyltransferase, and N-acetylglucosaminyltransferase. GT-8 enzymes contains a conserved DXD motif which is essential in the coordination of a catalytic divalent cation, most commonly Mn2+. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.75e-168 | 5 | 357 | 6 | 361 | |
| 2.38e-164 | 7 | 391 | 4 | 395 | |
| 7.14e-159 | 1 | 340 | 1 | 344 | |
| 1.04e-145 | 10 | 326 | 10 | 329 | |
| 1.48e-145 | 1 | 326 | 1 | 329 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.58e-10 | 7 | 298 | 6 | 258 | UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Platinum [Globisporangium ultimum],6MW8_A UDP-galactose:glucoside-Skp1 alpha-D-galactosyltransferase with bound UDP and Manganese [Globisporangium ultimum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.87e-20 | 5 | 270 | 16 | 253 | Galactinol synthase 1 OS=Solanum lycopersicum OX=4081 GN=GOLS1 PE=2 SV=1 |
|
| 1.28e-19 | 2 | 270 | 14 | 267 | Galactinol synthase 2 OS=Solanum lycopersicum OX=4081 GN=GOLS2 PE=2 SV=1 |
|
| 2.55e-19 | 2 | 292 | 24 | 296 | Galactinol synthase 1 OS=Arabidopsis thaliana OX=3702 GN=GOLS1 PE=1 SV=1 |
|
| 7.65e-19 | 6 | 270 | 21 | 266 | Galactinol synthase 1 OS=Ajuga reptans OX=38596 GN=GOLS1 PE=1 SV=1 |
|
| 2.67e-18 | 7 | 270 | 20 | 266 | Galactinol synthase 2 OS=Arabidopsis thaliana OX=3702 GN=GOLS2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000036 | 0.000002 |