You are browsing environment: FUNGIDB
CAZyme Information: OTA32993.1
You are here: Home > Sequence: OTA32993.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA32993.1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | Mannan endo-1,6-alpha-mannosidase [Source:UniProtKB/TrEMBL;Acc:A0A1Z5TAF3] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.101:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 74 | 311 | 3.8e-62 | 0.5865921787709497 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 6.90e-101 | 29 | 301 | 1 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 215151 | PLN02267 | 7.89e-32 | 335 | 523 | 30 | 224 | enoyl-CoA hydratase/isomerase family protein |
| 119339 | crotonase-like | 1.77e-31 | 331 | 492 | 26 | 191 | Crotonase/Enoyl-Coenzyme A (CoA) hydratase superfamily. This superfamily contains a diverse set of enzymes including enoyl-CoA hydratase, napthoate synthase, methylmalonyl-CoA decarboxylase, 3-hydoxybutyryl-CoA dehydratase, and dienoyl-CoA isomerase. Many of these play important roles in fatty acid metabolism. In addition to a conserved structural core and the formation of trimers (or dimers of trimers), a common feature in this superfamily is the stabilization of an enolate anion intermediate derived from an acyl-CoA substrate. This is accomplished by two conserved backbone NH groups in active sites that form an oxyanion hole. |
| 223955 | CaiD | 1.99e-20 | 331 | 546 | 32 | 246 | Enoyl-CoA hydratase/carnithine racemase [Lipid transport and metabolism]. |
| 395302 | ECH_1 | 9.26e-17 | 339 | 477 | 31 | 171 | Enoyl-CoA hydratase/isomerase. This family contains a diverse set of enzymes including: enoyl-CoA hydratase, napthoate synthase, carnitate racemase, 3-hydroxybutyryl-CoA dehydratase and dodecanoyl-CoA delta-isomerase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.93e-111 | 5 | 323 | 7 | 409 | |
| 1.34e-108 | 5 | 325 | 4 | 407 | |
| 1.34e-108 | 5 | 325 | 4 | 407 | |
| 1.34e-108 | 5 | 325 | 4 | 407 | |
| 2.41e-106 | 5 | 320 | 4 | 402 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.71e-50 | 74 | 323 | 171 | 414 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
|
| 4.30e-08 | 335 | 492 | 30 | 195 | Chain A, Enoyl-CoA Hydratase [Thermus thermophilus] |
|
| 1.20e-07 | 75 | 208 | 145 | 276 | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.56e-07 | 334 | 491 | 36 | 194 | Crystal structure of Enoyl-CoA hydratase, EchA3, from Mycobacterium ulcerans Agy99 [Mycobacterium ulcerans Agy99],6C7C_B Crystal structure of Enoyl-CoA hydratase, EchA3, from Mycobacterium ulcerans Agy99 [Mycobacterium ulcerans Agy99],6C7C_C Crystal structure of Enoyl-CoA hydratase, EchA3, from Mycobacterium ulcerans Agy99 [Mycobacterium ulcerans Agy99] |
|
| 2.05e-07 | 334 | 491 | 34 | 192 | Crystal structure of an enoyl-CoA hydratase (ECHA3) from Mycobacterium marinum [Mycobacterium marinum M] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.69e-75 | 74 | 308 | 148 | 379 | Putative mannan endo-1,6-alpha-mannosidase C1198.07c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1198.07c PE=3 SV=2 |
|
| 1.61e-72 | 74 | 309 | 155 | 389 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DCW1 PE=1 SV=1 |
|
| 7.52e-70 | 72 | 310 | 154 | 386 | Putative mannan endo-1,6-alpha-mannosidase C970.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC970.02 PE=3 SV=1 |
|
| 1.31e-67 | 14 | 322 | 13 | 391 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
|
| 2.76e-67 | 19 | 314 | 21 | 391 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DCW1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
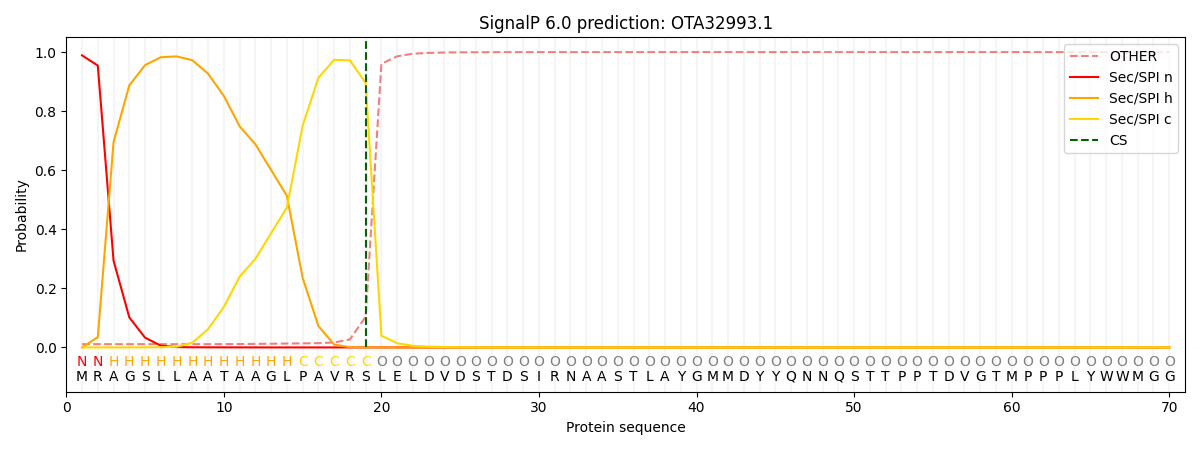
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.014401 | 0.985591 | CS pos: 19-20. Pr: 0.8958 |
