You are browsing environment: FUNGIDB
CAZyme Information: OTA32837.1
You are here: Home > Sequence: OTA32837.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
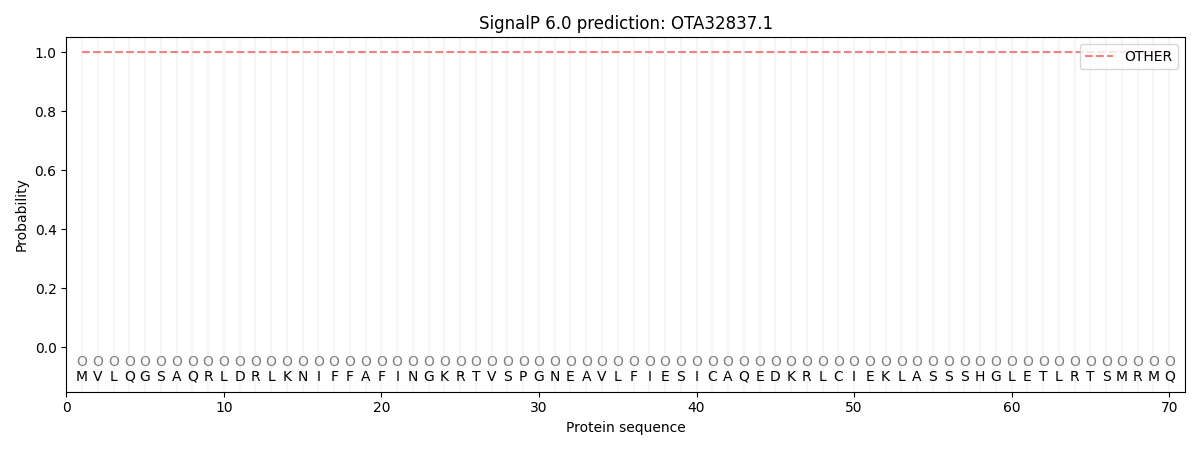
SignalP and Lipop annotations |
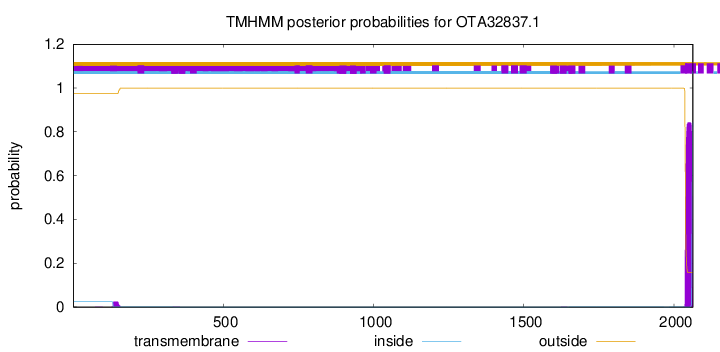
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA32837.1 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.58:29 | 3.2.1.21:6 | 3.2.1.75:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 1692 | 1940 | 8.5e-76 | 0.7722772277227723 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350694 | EEXXEc_NFX1 | 1.05e-61 | 465 | 820 | 1 | 178 | EEXXE-box helicase domain of NFX1. Human NFX1 protein was identified as a protein that represses class II MHC (major histocompatibility complex) gene expression. NFX1 binds a conserved cis-acting element, termed the X-box, in promoters of human class II MHC genes. The Cys-rich region contains several NFX1-type zinc finger domains. Frequently, a R3H domain is present in the C-terminus, and a RING finger domain and a PAM2 motif are present in the N-terminus. The lack of R3H and PAM2 motifs in the plant proteins indicates functional differences. Plant NFX1-like proteins are proposed to modulate growth and survival by coordinating reactive oxygen species, salicylic acid, further biotic stress and abscisic acid responses. A common feature of all members may be E3 ubiquitin ligase, due to the presence of a RING finger domain, as well as DNA binding. NFX1 is a member of the DEAD-like helicase superfamily, a diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. This domain contains the ATP-binding region. |
| 225344 | BglC | 3.28e-52 | 1641 | 1999 | 13 | 374 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 350195 | SF1_C_Upf1 | 1.34e-48 | 822 | 1036 | 1 | 183 | C-terminal helicase domain of Upf1-like family helicases. The Upf1-like helicase family includes UPF1, HELZ, Mov10L1, Aquarius, IGHMBP2 (SMUBP2), and similar proteins. They are DEAD-like helicases belonging to superfamily (SF)1, a diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. Similar to SF2 helicases, SF1 helicases do not form toroidal structures like SF3-6 helicases. Their helicase core consists of two similar protein domains that resemble the fold of the recombination protein RecA. This model describes the C-terminal domain, also called HelicC. |
| 404073 | AAA_12 | 1.07e-46 | 798 | 1017 | 1 | 196 | AAA domain. This family of domains contain a P-loop motif that is characteristic of the AAA superfamily. Many of the proteins in this family are conjugative transfer proteins. |
| 224037 | DNA2 | 8.51e-37 | 679 | 1020 | 433 | 732 | Superfamily I DNA and/or RNA helicase [Replication, recombination and repair]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.26e-127 | 1643 | 2034 | 40 | 456 | |
| 1.95e-116 | 1647 | 2047 | 26 | 397 | |
| 2.95e-69 | 1646 | 2009 | 73 | 454 | |
| 4.01e-69 | 1646 | 2009 | 73 | 454 | |
| 3.29e-67 | 1646 | 2009 | 73 | 454 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.20e-56 | 1646 | 2012 | 9 | 384 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 3.25e-55 | 1646 | 2012 | 9 | 384 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 3.80e-55 | 1646 | 2012 | 15 | 390 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
|
| 5.02e-55 | 1646 | 2012 | 14 | 389 | F144Y/F258Y Double Mutant of Exo-beta-1,3-glucanase from Candida albicans at 2 A [Candida albicans] |
|
| 9.49e-55 | 1646 | 2012 | 15 | 390 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.05e-84 | 203 | 1137 | 320 | 1340 | NFX1-type zinc finger-containing protein 1 OS=Mus musculus OX=10090 GN=Znfx1 PE=1 SV=3 |
|
| 2.74e-84 | 209 | 1137 | 333 | 1347 | NFX1-type zinc finger-containing protein 1 OS=Homo sapiens OX=9606 GN=ZNFX1 PE=1 SV=2 |
|
| 4.03e-61 | 1641 | 2018 | 34 | 405 | Glucan 1,3-beta-glucosidase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=exg1 PE=2 SV=1 |
|
| 2.37e-60 | 1646 | 1989 | 42 | 386 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
|
| 8.04e-60 | 1646 | 2012 | 42 | 408 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000064 | 0.000000 |