You are browsing environment: FUNGIDB
CAZyme Information: OTA31852.1
You are here: Home > Sequence: OTA31852.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA31852.1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH92 | 511 | 1024 | 1.4e-158 | 0.9898167006109979 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400362 | Glyco_hydro_92 | 0.0 | 541 | 1023 | 2 | 464 | Glycosyl hydrolase family 92. Members of this family are alpha-1,2-mannosidases, enzymes which remove alpha-1,2-linked mannose residues from Man(9)(GlcNAc)(2) by hydrolysis. They are critical for the maturation of N-linked oligosaccharides and ER-associated degradation. |
| 226067 | COG3537 | 3.47e-129 | 272 | 1049 | 33 | 767 | Putative alpha-1,2-mannosidase [Carbohydrate transport and metabolism]. |
| 235797 | PRK06411 | 5.44e-102 | 76 | 229 | 14 | 164 | NADH-quinone oxidoreductase subunit NuoB. |
| 223454 | NuoB | 3.94e-94 | 65 | 228 | 1 | 164 | NADH:ubiquinone oxidoreductase 20 kD subunit (chhain B) or related Fe-S oxidoreductase [Energy production and conversion]. |
| 273896 | nuoB_fam | 9.60e-92 | 87 | 225 | 4 | 143 | NADH-quinone oxidoreductase, B subunit. This model describes the B chain of complexes that resemble NADH-quinone oxidoreductases. The electron acceptor is a quinone, ubiquinone, in mitochondria and most bacteria, including Escherichia coli, where the recommended gene symbol is nuoB. The quinone is plastoquinone in Synechocystis (where the chain is designated K) and in chloroplast, where NADH may be replaced by NADPH. In the methanogenic archaeal genus Methanosarcina, NADH is replaced by F420H2. [Energy metabolism, Electron transport] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 249 | 1049 | 2 | 804 | |
| 0.0 | 276 | 1042 | 61 | 827 | |
| 0.0 | 276 | 1042 | 31 | 808 | |
| 0.0 | 276 | 1042 | 31 | 808 | |
| 0.0 | 273 | 1043 | 29 | 810 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.97e-94 | 57 | 230 | 5 | 177 | Chain K, Subunit NUKM of protein NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica] |
|
| 1.01e-93 | 57 | 230 | 32 | 204 | Chain K, PSST SUBUNIT (NUKM) [Yarrowia lipolytica],6RFQ_K Chain K, Subunit NUKM of NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica],6RFR_K Chain K, Subunit NUKM of NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica],6RFS_K Cryo-EM structure of a respiratory complex I mutant lacking NDUFS4 [Yarrowia lipolytica],6Y79_K Cryo-EM structure of a respiratory complex I F89A mutant [Yarrowia lipolytica],6YJ4_B Structure of Yarrowia lipolytica complex I at 2.7 A [Yarrowia lipolytica],7B0N_B Chain B, Subunit NUKM of NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica],7O6Y_K Chain K, Subunit NUKM of NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica],7O71_K Chain K, Subunit NUKM of NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica] |
|
| 8.70e-89 | 85 | 230 | 1 | 146 | Chain K, Subunit NUKM of protein NADH:Ubiquinone Oxidoreductase (Complex I) [Yarrowia lipolytica] |
|
| 6.59e-82 | 73 | 228 | 21 | 171 | Structure of mammalian respiratory Complex I, class1 [Bos taurus],5LDX_B Structure of mammalian respiratory Complex I, class3. [Bos taurus],5LNK_6 Chain 6, Mitochondrial complex I, PSST subunit [Ovis aries],5O31_B Mitochondrial complex I in the deactive state [Bos taurus],6Q9D_S7 Chain S7, NADH:ubiquinone oxidoreductase core subunit S7 [Ovis aries],6QA9_S7 Chain S7, NDUFS7 [Ovis aries],6QBX_S7 Chain S7, NDUFS7 [Ovis aries],6QC2_S7 Chain S7, NDUFS7 [Ovis aries],6QC3_S7 Chain S7, NDUFB1 [Ovis aries],6QC4_S7 Chain S7, NDUFS7 [Ovis aries],6QC5_S7 Chain S7, NADH:ubiquinone oxidoreductase core subunit S7,NDUFS7 [Ovis aries],6QC6_S7 Chain S7, NADH:ubiquinone oxidoreductase core subunit S7 [Ovis aries],6QC7_S7 Chain S7, NDUFS7 [Ovis aries],6QC8_S7 Chain S7, NADH:ubiquinone oxidoreductase core subunit S7 [Ovis aries],6QC9_S7 Chain S7, NDUFS7 [Ovis aries],6QCA_S7 Chain S7, NDUFS7 [Ovis aries],6QCF_S7 Chain S7, NDUFS7 [Ovis aries],7DGQ_D Chain D, NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Bos taurus],7DGR_D Chain D, NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Bos taurus],7DGS_D Chain D, NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Bos taurus],7DGZ_D Chain D, NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Bos taurus],7DH0_D Chain D, NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Bos taurus],7DKF_D2 Chain D2, NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial [Bos taurus] |
|
| 8.18e-82 | 84 | 228 | 1 | 145 | Structure of mammalian respiratory Complex I, class2 [Bos taurus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 250 | 1040 | 3 | 758 | Uncharacterized secreted glycosidase ARB_07629 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07629 PE=1 SV=1 |
|
| 5.25e-110 | 1 | 230 | 1 | 220 | NADH-ubiquinone oxidoreductase 19.3 kDa subunit, mitochondrial OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=G17A4.170 PE=2 SV=1 |
|
| 6.12e-81 | 78 | 227 | 2 | 153 | NADH-quinone oxidoreductase subunit B 2 OS=Acidithiobacillus ferrooxidans (strain ATCC 53993 / BNL-5-31) OX=380394 GN=nuoB2 PE=3 SV=1 |
|
| 7.75e-81 | 24 | 228 | 12 | 205 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial OS=Gorilla gorilla gorilla OX=9595 GN=NDUFS7 PE=2 SV=1 |
|
| 7.75e-81 | 24 | 228 | 12 | 205 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial OS=Pongo pygmaeus OX=9600 GN=NDUFS7 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
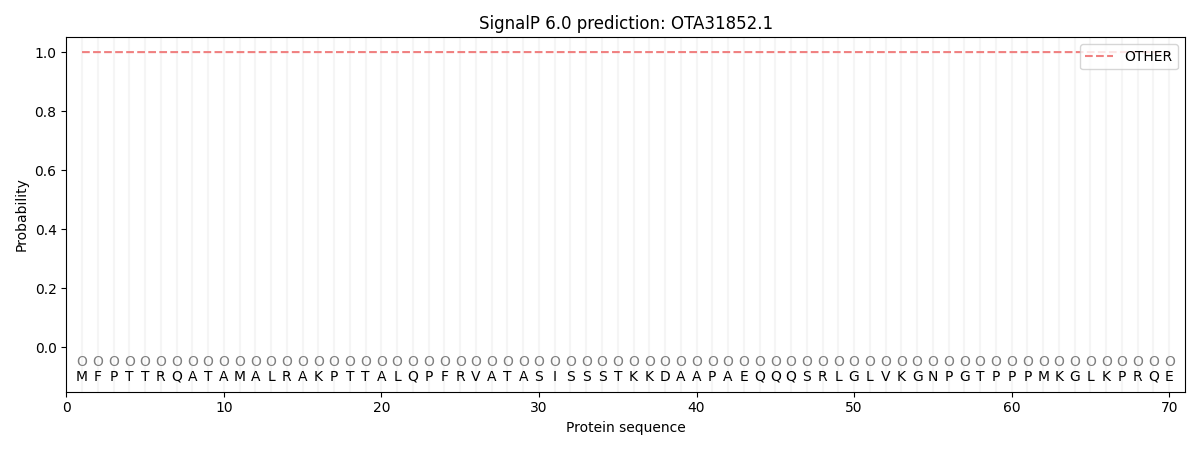
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000033 | 0.000000 |
