You are browsing environment: FUNGIDB
CAZyme Information: OTA31082.1
You are here: Home > Sequence: OTA31082.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
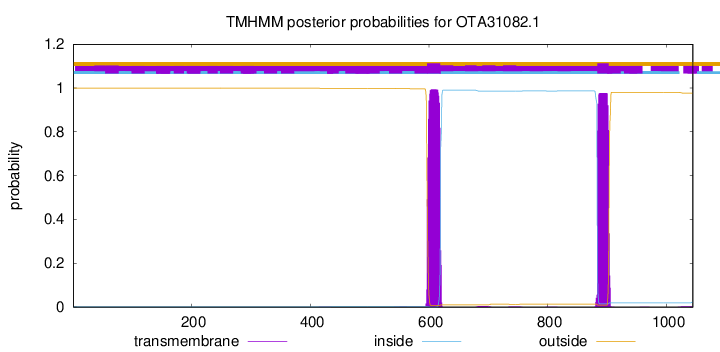
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA31082.1 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 677 | 1040 | 3.6e-42 | 0.8575418994413407 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 213362 | TmCorA-like_1 | 3.78e-25 | 198 | 617 | 3 | 252 | Thermotoga maritima CorA_like subfamily. This subfamily belongs to the Thermotoga maritima CorA (TmCorA)-family of the MIT superfamily of essential membrane proteins involved in transporting divalent cations (uptake or efflux) across membranes. Members of this subfamily are found in all three kingdoms of life. It is functionally diverse subfamily, in addition to the CorA Co2+ transporter from the hyperthermophilic Thermotoga maritima, it includes Methanosarcina mazei CorA which may be involved in transport of copper and/or other divalent metal ions. Thermotoga maritima CorA forms funnel-shaped homopentamers, the tip of the funnel is formed from two C-terminal transmembrane (TM) helices from each monomer, and the large opening of the funnel from the N-terminal cytoplasmic domains. The GMN signature motif of the MIT superfamily occurs just after TM1, mutation within this motif is known to abolish Mg2+ transport by a related protein, Saccharomyces cerevisiae Alr1p. Natural variants in this signature sequence may be associated with the transport of different divalent cations. The functional diversity of the MIT superfamily may also be due to minor structural differences regulating gating, substrate selection, and transport. |
| 397638 | Glyco_hydro_76 | 2.19e-20 | 686 | 1033 | 35 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| 213356 | TmCorA-like | 2.38e-14 | 199 | 619 | 1 | 249 | Thermotoga maritima CorA-like family. This family belongs to the MIT superfamily of essential membrane proteins involved in transporting divalent cations (uptake or efflux) across membranes. Members of the Thermotoga maritima CorA_like family are found in all three kingdoms of life. It is a functionally diverse family, in addition to the CorA Co2+ transporter from the hyperthermophilic Thermotoga maritima, it includes three Saccharomyces cerevisiae members: two plasma membrane proteins, the Mg2+ transporter Alr1p/Swc3p and the putative Mg2+ transporter, Alr2p, and the vacuole membrane protein Mnr2p, a putative Mg2+ transporter. Thermotoga maritima CorA forms funnel-shaped homopentamers, the tip of the funnel is formed from two C-terminal transmembrane (TM) helices from each monomer, and the large opening of the funnel from the N-terminal cytoplasmic domains. The GMN signature motif of the MIT superfamily occurs just after TM1, mutation within this motif is known to abolish Mg2+ transport by Alr1p. Natural variants in this signature sequence may be associated with the transport of different divalent cations. The functional diversity of the MIT superfamily may also be due to minor structural differences regulating gating, substrate selection, and transport. |
| 223671 | CorA | 2.11e-09 | 355 | 619 | 80 | 282 | Mg2+ and Co2+ transporter CorA [Inorganic ion transport and metabolism]. |
| 396223 | CorA | 2.96e-09 | 551 | 619 | 186 | 254 | CorA-like Mg2+ transporter protein. The CorA transport system is the primary Mg2+ influx system of Salmonella typhimurium and Escherichia coli. CorA is virtually ubiquitous in the Bacteria and Archaea. There are also eukaryotic relatives of this protein. The family includes the MRS2 protein from yeast that is thought to be an RNA splicing protein. However its membership of this family suggests that its effect on splicing is due to altered magnesium levels in the cell. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.44e-158 | 633 | 1043 | 5 | 406 | |
| 1.55e-26 | 655 | 1043 | 34 | 395 | |
| 3.73e-26 | 663 | 1043 | 41 | 394 | |
| 1.24e-25 | 663 | 1043 | 41 | 395 | |
| 9.96e-25 | 678 | 1043 | 57 | 395 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.37e-06 | 145 | 625 | 7 | 317 | Cryo-EM structure of the magnesium channel CorA in the closed symmetric magnesium-bound state [Thermotoga maritima],3JCF_B Cryo-EM structure of the magnesium channel CorA in the closed symmetric magnesium-bound state [Thermotoga maritima],3JCF_C Cryo-EM structure of the magnesium channel CorA in the closed symmetric magnesium-bound state [Thermotoga maritima],3JCF_D Cryo-EM structure of the magnesium channel CorA in the closed symmetric magnesium-bound state [Thermotoga maritima],3JCF_E Cryo-EM structure of the magnesium channel CorA in the closed symmetric magnesium-bound state [Thermotoga maritima],3JCG_A Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state I [Thermotoga maritima],3JCG_B Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state I [Thermotoga maritima],3JCG_C Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state I [Thermotoga maritima],3JCG_D Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state I [Thermotoga maritima],3JCG_E Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state I [Thermotoga maritima],3JCH_A Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state II [Thermotoga maritima],3JCH_B Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state II [Thermotoga maritima],3JCH_C Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state II [Thermotoga maritima],3JCH_D Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state II [Thermotoga maritima],3JCH_E Cryo-EM structure of the magnesium channel CorA in the magnesium-free, asymmetric open state II [Thermotoga maritima],4I0U_A Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_B Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_C Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_D Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_E Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_F Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_G Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_H Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_I Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8],4I0U_J Improved structure of Thermotoga maritima CorA at 2.7 A resolution [Thermotoga maritima MSB8] |
|
| 1.39e-06 | 145 | 625 | 10 | 320 | Crystal structure of the CorA Mg2+ transporter [Thermotoga maritima],2BBJ_B Crystal structure of the CorA Mg2+ transporter [Thermotoga maritima],2BBJ_D Crystal structure of the CorA Mg2+ transporter [Thermotoga maritima],2BBJ_E Crystal structure of the CorA Mg2+ transporter [Thermotoga maritima],2BBJ_F Crystal structure of the CorA Mg2+ transporter [Thermotoga maritima],2HN2_A Crystal structure of the CorA Mg2+ transporter homologue from T. maritima in complex with divalent cations [Thermotoga maritima],2HN2_B Crystal structure of the CorA Mg2+ transporter homologue from T. maritima in complex with divalent cations [Thermotoga maritima],2HN2_C Crystal structure of the CorA Mg2+ transporter homologue from T. maritima in complex with divalent cations [Thermotoga maritima],2HN2_D Crystal structure of the CorA Mg2+ transporter homologue from T. maritima in complex with divalent cations [Thermotoga maritima],2HN2_E Crystal structure of the CorA Mg2+ transporter homologue from T. maritima in complex with divalent cations [Thermotoga maritima] |
|
| 1.44e-06 | 145 | 625 | 19 | 329 | Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_B Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_C Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_D Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_E Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_F Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_G Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_H Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_I Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima],2IUB_J Crystal structure of a divalent metal ion transporter CorA at 2.9 A resolution. [Thermotoga maritima] |
|
| 4.78e-06 | 728 | 1037 | 131 | 387 | Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.06e-06 | 145 | 625 | 7 | 317 | Cobalt/magnesium transport protein CorA OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=corA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000061 | 0.000000 |