You are browsing environment: FUNGIDB
CAZyme Information: OTA26897.1
You are here: Home > Sequence: OTA26897.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA26897.1 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | 1,4-alpha-D-glucan glucohydrolase [Source:UniProtKB/TrEMBL;Acc:A0A1Z5SYW0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH15 | 224 | 648 | 2.2e-72 | 0.9473684210526315 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395586 | Glyco_hydro_15 | 3.28e-92 | 216 | 649 | 10 | 417 | Glycosyl hydrolases family 15. In higher organisms this family is represented by phosphorylase kinase subunits. |
| 225922 | SGA1 | 1.81e-16 | 227 | 658 | 262 | 611 | Glucoamylase (glucan-1,4-alpha-glucosidase), GH15 family [Carbohydrate transport and metabolism]. |
| 273702 | oligosac_amyl | 4.23e-12 | 186 | 645 | 226 | 607 | oligosaccharide amylase. The name of this type of amylase is based on the characterization of an glucoamylase family enzyme from Thermoactinomyces vulgaris. The T. vulgaris enzyme was expressed in E. coli and, like other glucoamylases, it releases beta-D-glucose from starch. However, unlike previously characterized glucoamylases, this T. vulgaris amylase hydrolyzes maltooligosaccharides (maltotetraose, maltose) more efficiently than starch (1), indicating this enzyme belongs to a class of glucoamylase-type enzymes with oligosaccharide-metabolizing activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.32e-254 | 30 | 667 | 1 | 624 | |
| 2.60e-234 | 36 | 668 | 11 | 630 | |
| 2.59e-211 | 52 | 667 | 80 | 674 | |
| 3.62e-105 | 160 | 661 | 17 | 484 | |
| 3.62e-105 | 160 | 661 | 17 | 484 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.71e-66 | 202 | 647 | 7 | 425 | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol [Aspergillus niger] |
|
| 6.90e-65 | 202 | 647 | 7 | 425 | Structure of the catalytic domain of Aspergillus niger Glucoamylase [Aspergillus niger] |
|
| 1.03e-64 | 202 | 647 | 7 | 424 | Refined structure for the complex of acarbose with glucoamylase from Aspergillus awamori var. x100 to 2.4 angstroms resolution [Aspergillus awamori],1DOG_A REFINED STRUCTURE FOR THE COMPLEX OF 1-DEOXYNOJIRIMYCIN WITH GLUCOAMYLASE FROM (ASPERGILLUS AWAMORI) VAR. X100 TO 2.4 ANGSTROMS RESOLUTION [Aspergillus awamori],1GLM_A REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 [Aspergillus awamori],3GLY_A REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 [Aspergillus awamori] |
|
| 1.06e-64 | 202 | 647 | 7 | 424 | GLUCOAMYLASE-471 COMPLEXED WITH ACARBOSE [Aspergillus awamori] |
|
| 1.08e-64 | 202 | 647 | 7 | 424 | GLUCOAMYLASE-471 COMPLEXED WITH D-GLUCO-DIHYDROACARBOSE [Aspergillus awamori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.35e-255 | 30 | 667 | 1 | 624 | Glucoamylase OS=Blastobotrys adeninivorans OX=409370 GN=GAA PE=3 SV=1 |
|
| 1.68e-94 | 59 | 654 | 26 | 601 | Glucoamylase 1 OS=Rhizopus oryzae OX=64495 PE=1 SV=2 |
|
| 1.12e-69 | 190 | 651 | 34 | 445 | Glucoamylase amyD OS=Rhizopus delemar (strain RA 99-880 / ATCC MYA-4621 / FGSC 9543 / NRRL 43880) OX=246409 GN=amyD PE=1 SV=1 |
|
| 3.07e-69 | 202 | 655 | 29 | 455 | Glucoamylase ARB_02327-1 OS=Schizophyllum commune (strain H4-8 / FGSC 9210) OX=578458 GN=SCHCODRAFT_57589 PE=1 SV=1 |
|
| 4.70e-66 | 202 | 647 | 34 | 451 | Glucoamylase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=glaA PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
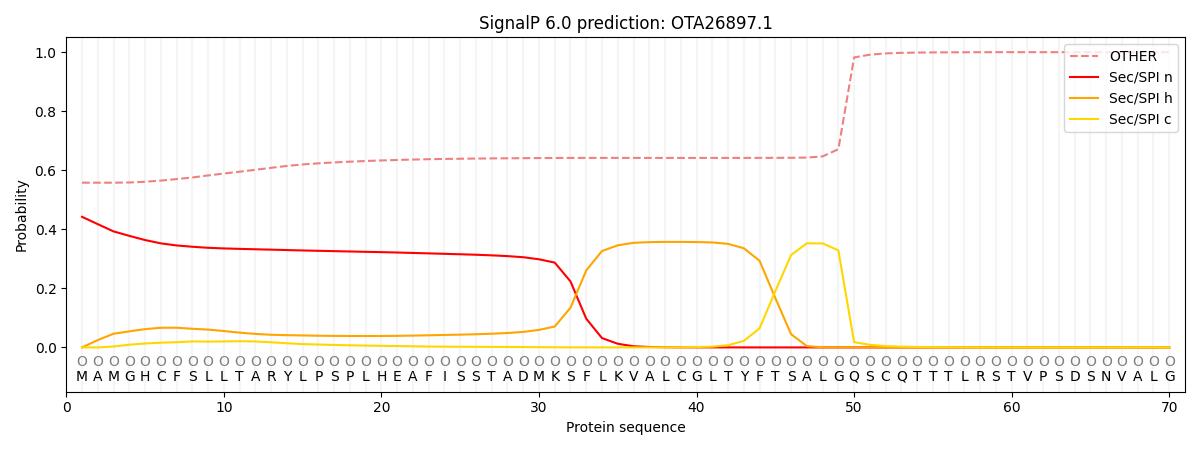
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.565627 | 0.434374 |
