You are browsing environment: FUNGIDB
CAZyme Information: OTA24895.1
You are here: Home > Sequence: OTA24895.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA24895.1 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH125 | 69 | 482 | 2.6e-157 | 0.9701492537313433 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 399660 | Glyco_hydro_125 | 0.0 | 69 | 481 | 1 | 404 | Metal-independent alpha-mannosidase (GH125). This family, which contains bacterial and fungal glycoside hydrolases, is also known as GH125. They function as metal-independent alpha-mannosidases, with specificity for alpha-1,6-linked non-reducing terminal mannose residues. Structurally this family is part of the 6 hairpin glycosidase superfamily. |
| 226068 | COG3538 | 6.99e-137 | 47 | 481 | 1 | 408 | Meiotically up-regulated gene 157 (Mug157) protein (function unknown) [Function unknown]. |
| 187632 | 17beta-HSD-like_SDR_c | 5.92e-90 | 481 | 728 | 2 | 248 | 17beta hydroxysteroid dehydrogenase-like, classical (c) SDRs. 17beta-hydroxysteroid dehydrogenases are a group of isozymes that catalyze activation and inactivation of estrogen and androgens. SDRs are a functionally diverse family of oxidoreductases that have a single domain with a structurally conserved Rossmann fold (alpha/beta folding pattern with a central beta-sheet), an NAD(P)(H)-binding region, and a structurally diverse C-terminal region. Classical SDRs are typically about 250 residues long, while extended SDRs are approximately 350 residues. Sequence identity between different SDR enzymes are typically in the 15-30% range, but the enzymes share the Rossmann fold NAD-binding motif and characteristic NAD-binding and catalytic sequence patterns. These enzymes catalyze a wide range of activities including the metabolism of steroids, cofactors, carbohydrates, lipids, aromatic compounds, and amino acids, and act in redox sensing. Classical SDRs have an TGXXX[AG]XG cofactor binding motif and a YXXXK active site motif, with the Tyr residue of the active site motif serving as a critical catalytic residue (Tyr-151, human 15-hydroxyprostaglandin dehydrogenase (15-PGDH) numbering). In addition to the Tyr and Lys, there is often an upstream Ser (Ser-138, 15-PGDH numbering) and/or an Asn (Asn-107, 15-PGDH numbering) contributing to the active site; while substrate binding is in the C-terminal region, which determines specificity. The standard reaction mechanism is a 4-pro-S hydride transfer and proton relay involving the conserved Tyr and Lys, a water molecule stabilized by Asn, and nicotinamide. Extended SDRs have additional elements in the C-terminal region, and typically have a TGXXGXXG cofactor binding motif. Complex (multidomain) SDRs such as ketoreductase domains of fatty acid synthase have a GGXGXXG NAD(P)-binding motif and an altered active site motif (YXXXN). Fungal type ketoacyl reductases have a TGXXXGX(1-2)G NAD(P)-binding motif. Some atypical SDRs have lost catalytic activity and/or have an unusual NAD(P)-binding motif and missing or unusual active site residues. Reactions catalyzed within the SDR family include isomerization, decarboxylation, epimerization, C=N bond reduction, dehydratase activity, dehalogenation, Enoyl-CoA reduction, and carbonyl-alcohol oxidoreduction. |
| 180446 | PRK06180 | 9.81e-86 | 482 | 753 | 7 | 276 | short chain dehydrogenase; Provisional |
| 181334 | PRK08263 | 3.44e-76 | 477 | 753 | 3 | 272 | short chain dehydrogenase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.25e-247 | 7 | 486 | 6 | 494 | |
| 1.25e-247 | 7 | 486 | 6 | 494 | |
| 1.71e-234 | 2 | 481 | 7 | 487 | |
| 1.71e-234 | 2 | 481 | 7 | 487 | |
| 1.71e-234 | 2 | 481 | 7 | 487 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.23e-80 | 38 | 504 | 38 | 472 | Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 [Bacteroides thetaiotaomicron VPI-5482],2P0V_B Crystal structure of BT3781 protein from Bacteroides thetaiotaomicron, Northeast Structural Genomics Target BtR58 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 2.38e-79 | 43 | 504 | 21 | 454 | Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03347) from bacteroides ovatus at 1.60 a resolution [Bacteroides ovatus ATCC 8483],3P2C_B Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03347) from bacteroides ovatus at 1.60 a resolution [Bacteroides ovatus ATCC 8483] |
|
| 3.14e-79 | 38 | 504 | 18 | 452 | Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03626) from bacteroides ovatus at 1.70 a resolution [Bacteroides ovatus ATCC 8483],3ON6_B Crystal structure of an exo-alpha-1,6-mannosidase (bacova_03626) from bacteroides ovatus at 1.70 a resolution [Bacteroides ovatus ATCC 8483] |
|
| 5.52e-72 | 63 | 496 | 19 | 423 | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure [Clostridium perfringens],3QT9_A Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Clostridium perfringens CPE0426 complexed with alpha-1,6-linked 1-thio-alpha-mannobiose [Clostridium perfringens] |
|
| 6.87e-72 | 63 | 501 | 19 | 429 | Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole [Clostridium perfringens str. 13],6RQK_B Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole [Clostridium perfringens str. 13] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.63e-123 | 18 | 481 | 27 | 482 | Meiotically up-regulated gene 157 protein OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mug157 PE=1 SV=1 |
|
| 1.09e-53 | 484 | 761 | 10 | 281 | Uncharacterized oxidoreductase C162.03 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC162.03 PE=3 SV=1 |
|
| 8.77e-40 | 477 | 730 | 2 | 253 | Oxidoreductase BOA17 OS=Botryotinia fuckeliana (strain B05.10) OX=332648 GN=BOA17 PE=2 SV=1 |
|
| 1.77e-36 | 483 | 736 | 18 | 280 | Oxidoreductase calM OS=Penicillium decumbens OX=69771 GN=calM PE=1 SV=1 |
|
| 1.55e-25 | 484 | 724 | 8 | 252 | Uncharacterized oxidoreductase YusZ OS=Bacillus subtilis (strain 168) OX=224308 GN=yusZ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
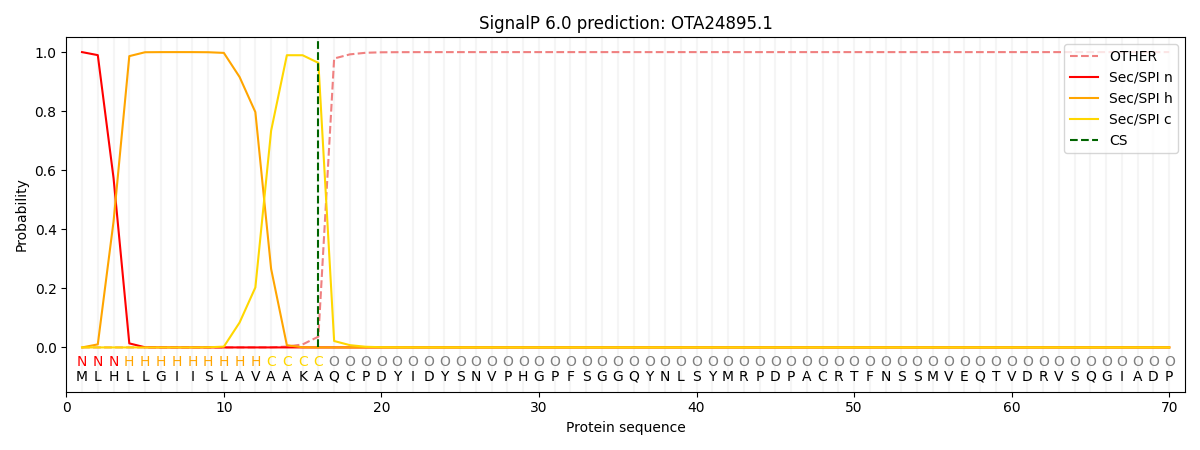
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000212 | 0.999770 | CS pos: 16-17. Pr: 0.9635 |
