You are browsing environment: FUNGIDB
CAZyme Information: OTA24810.1
You are here: Home > Sequence: OTA24810.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA24810.1 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 32 | 444 | 5e-163 | 0.9850374064837906 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 1.39e-16 | 1 | 449 | 14 | 399 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| 402070 | Lactonase | 0.004 | 56 | 120 | 193 | 262 | Lactonase, 7-bladed beta-propeller. This entry contains bacterial 6-phosphogluconolactonases (6PGL)YbhE-type (EC:3.1.1.31) which hydrolyze 6-phosphogluconolactone to 6-phosphogluconate. The entry also contains the fungal muconate lactonising enzyme carboxy-cis,cis-muconate cyclase (EC:5.5.1.5) and muconate cycloisomerase (EC:5.5.1.1), which convert cis,cis-muconates to muconolactones and vice versa as part of the microbial beta-ketoadipate pathway. Structures of proteins in this family have revealed a 7-bladed beta-propeller fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.21e-172 | 13 | 456 | 18 | 440 | |
| 1.93e-153 | 10 | 450 | 12 | 430 | |
| 8.89e-148 | 10 | 461 | 15 | 441 | |
| 8.89e-148 | 10 | 461 | 15 | 441 | |
| 8.89e-148 | 10 | 461 | 15 | 441 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.96e-126 | 35 | 447 | 8 | 403 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 3.06e-126 | 35 | 447 | 9 | 404 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 3.17e-126 | 35 | 447 | 10 | 405 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 5.21e-72 | 30 | 446 | 1 | 410 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.11e-10 | 37 | 448 | 71 | 445 | L-sorbosone dehydrogenase OS=Gluconacetobacter liquefaciens OX=89584 PE=4 SV=1 |
|
| 5.05e-06 | 227 | 388 | 193 | 326 | Uncharacterized protein BB_0024 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0024 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
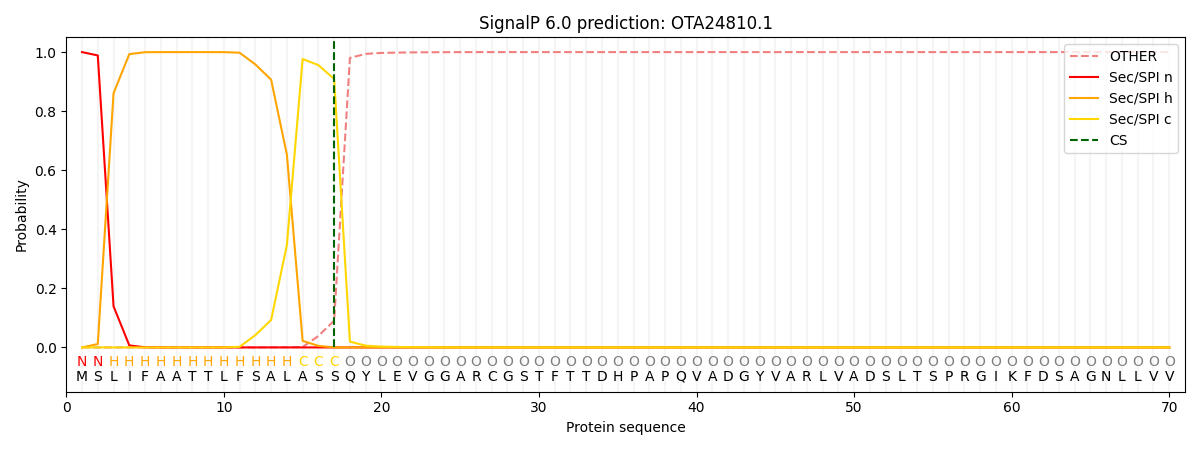
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000323 | 0.999661 | CS pos: 17-18. Pr: 0.9092 |
