You are browsing environment: FUNGIDB
CAZyme Information: OTA24504.1
You are here: Home > Sequence: OTA24504.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA24504.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 6650; End:10242 Strand: + | |||||||||||
Full Sequence Download help
| MSPRSTKRER EPLPSCSSTI IPNRMTCSRK LFIAFAASTV HVVQAQQLKD GHPAFMSYPG | 60 |
| INNACETALN TTVDCPAFLQ RVSVNNAILD LDQVQALCTG SCVTDLQSAR SDIASACTAD | 120 |
| TDVIVYDNVA YPATFIADQY LFTTEVSCLR DTYTGEYCDP KFLVWSNQSF MTTNQSCSNC | 180 |
| WLNVQALQLG NPLAYDDSLA SNFAELKASC NAKSYTYATP TVYGINATET TDTDGAKFTK | 240 |
| PATCTGSYVL QPGDDCNSVA KKMGVSTYSM LVSNGLDLYC QNFEAAVNSS ASLCTPPTCK | 300 |
| IYEWNFFDEC NDVANQNDVS VAHFLSWNPN FDSICRNSII FAGYQVCVSA PGGTVESGAS | 360 |
| TNLSVSPGAS SAVPAPTNSF PETKECGGWY TVQAGDDCAK LSVAFSMTIS DFLFLNPEIY | 420 |
| TNCTNLLLGV SYCVFPVGDI ASYSGYPAPT TLSITVPPAT FSSVNTAVPS ATNHPDPDFV | 480 |
| FQNLPKAPGT ADDCTFYADH NDNTRDADSN SCTAVASAWD IELEDLLDWN PSLSTDQEAC | 540 |
| ATQPGRSYCV RKSTKVVYAP TGDECIAVNA TEIPEGTNAD CTCFTEVSGY EGTVGLDCKA | 600 |
| IADDSSITLV VLQDLNPWLA SDCDNALFAG LGEWDHRAVC IGTNATSSSS STRSSSSGSK | 660 |
| ITKAMGPTRT GTPAGCRRFY TVQSGDGCAA IEETFDITFA QLYKWNPTIG EDCANLWLGY | 720 |
| AYCVDASISA TITTSSPTRS TPSVTTPTPT QEGMTKSCND FYKVKSGDGC YDIASHYGIS | 780 |
| LEDFYTYNPA VGDDCSKLYP DNYVCVGVTS SGSCKIDVKF NTAHSTEWGE SVWAVGSIPE | 840 |
| LGSWDVNEAL MLTGSSGADG STNWQATAEL PADTQVSYKF VKVQTDGTPV WEQDPNRSFL | 900 |
| TSSCGGSAMQ EGGSWQGSST GSTAPSTTSP IVPTCTSLDV VFEVLAQTTY GESVYVIGSV | 960 |
| PALGEWSTDA AVVLAADQYT QARPLWRGTI SLAVGQDVQF KFIKINLDGT YTWEADPNRI | 1020 |
| VRIPTDCSAT LTQSGTFQQ | 1039 |
Enzyme Prediction help
| EC | - | 3.2.1.3:71 | 3.2.1.1:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM20 | 816 | 905 | 9.7e-24 | 0.9555555555555556 |
| CBM20 | 940 | 1028 | 1.9e-23 | 0.9444444444444444 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 99886 | CBM20_glucoamylase | 1.36e-36 | 933 | 1036 | 2 | 105 | Glucoamylase (glucan1,4-alpha-glucosidase), C-terminal CBM20 (carbohydrate-binding module, family 20) domain. Glucoamylases are inverting, exo-acting starch hydrolases that hydrolyze starch and related polysaccharides by releasing the nonreducing end glucose. They are mainly active on alpha-1,4-glycosidic bonds but also have some activity towards 1,6-glycosidic bonds occurring in natural oligosaccharides. The ability of glucoamylases to cleave 1-6-glycosidic binds is called "debranching activity" and is of importance in industrial applications, where complete degradation of starch to glucose is needed. Most glucoamylases are multidomain proteins containing an N-terminal catalytic domain, a C-terminal CBM20 domain, and a highly O-glycosylated linker region that connects the two. The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 395557 | CBM_20 | 6.36e-32 | 940 | 1032 | 3 | 94 | Starch binding domain. |
| 99883 | CBM20_alpha_amylase | 1.04e-26 | 940 | 1034 | 3 | 93 | Alpha-amylase, C-terminal CBM20 (carbohydrate-binding module, family 20) domain. This domain is found in several bacterial and fungal alpha-amylases including the maltopentaose-forming amylases (G5-amylases). Most alpha-amylases have, in addition to the C-terminal CBM20 domain, an N-terminal catalytic domain belonging to glycosyl hydrolase family 13, which hydrolyzes internal alpha-1,4-glucosidic bonds in starch and related saccharides, yielding maltotriose and maltose. Two types of soluble substrates are used by alpha-amylases including long substrates (e.g. amylose) and short substrates (e.g. maltodextrins or maltooligosaccharides). The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 119437 | CBM20 | 1.06e-24 | 940 | 1032 | 2 | 93 | The family 20 carbohydrate-binding module (CBM20), also known as the starch-binding domain, is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
| 99886 | CBM20_glucoamylase | 2.24e-24 | 818 | 915 | 9 | 106 | Glucoamylase (glucan1,4-alpha-glucosidase), C-terminal CBM20 (carbohydrate-binding module, family 20) domain. Glucoamylases are inverting, exo-acting starch hydrolases that hydrolyze starch and related polysaccharides by releasing the nonreducing end glucose. They are mainly active on alpha-1,4-glycosidic bonds but also have some activity towards 1,6-glycosidic bonds occurring in natural oligosaccharides. The ability of glucoamylases to cleave 1-6-glycosidic binds is called "debranching activity" and is of importance in industrial applications, where complete degradation of starch to glucose is needed. Most glucoamylases are multidomain proteins containing an N-terminal catalytic domain, a C-terminal CBM20 domain, and a highly O-glycosylated linker region that connects the two. The CBM20 domain is found in a large number of starch degrading enzymes including alpha-amylase, beta-amylase, glucoamylase, and CGTase (cyclodextrin glucanotransferase). CBM20 is also present in proteins that have a regulatory role in starch metabolism in plants (e.g. alpha-amylase) or glycogen metabolism in mammals (e.g. laforin). CBM20 folds as an antiparallel beta-barrel structure with two starch binding sites. These two sites are thought to differ functionally with site 1 acting as the initial starch recognition site and site 2 involved in the specific recognition of appropriate regions of starch. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW40759.1|CBM50 | 1.39e-135 | 28 | 727 | 3 | 743 |
| UNI22107.1|CBM50 | 6.17e-134 | 31 | 724 | 4 | 686 |
| ATY65640.1|CBM50 | 5.04e-129 | 31 | 807 | 14 | 779 |
| UKZ93577.1|CBM50 | 7.20e-121 | 35 | 641 | 11 | 601 |
| SMQ53700.1|CBM50 | 4.76e-119 | 33 | 461 | 10 | 441 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1AC0_A | 9.61e-24 | 934 | 1038 | 3 | 108 | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, MINIMIZED AVERAGE STRUCTURE [Aspergillus niger],1ACZ_A GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, 5 STRUCTURES [Aspergillus niger],1KUL_A Chain A, GLUCOAMYLASE [Aspergillus niger],1KUM_A Chain A, GLUCOAMYLASE [Aspergillus niger],5GHL_A Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger],5GHL_B Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger],5GHL_C Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger],5GHL_D Crystal structure Analysis of the starch-binding domain of glucoamylase from Aspergillus niger [Aspergillus niger] |
| 6FRV_A | 1.48e-19 | 942 | 1038 | 519 | 616 | Structure of the catalytic domain of Aspergillus niger Glucoamylase [Aspergillus niger] |
| 2VN4_A | 6.94e-18 | 889 | 1038 | 439 | 598 | Chain A, GLUCOAMYLASE [Trichoderma reesei],2VN7_A Chain A, GLUCOAMYLASE [Trichoderma reesei] |
| 6FHV_A | 1.60e-11 | 932 | 1027 | 486 | 582 | Crystal structure of Penicillium oxalicum Glucoamylase [Penicillium oxalicum 114-2] |
| 6FHW_A | 8.61e-11 | 811 | 917 | 502 | 608 | Chain A, Glucoamylase P [Amorphotheca resinae],6FHW_B Chain B, Glucoamylase P [Amorphotheca resinae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|D4AVW3|LYSM5_ARTBC | 5.47e-36 | 335 | 724 | 211 | 534 | LysM domain-containing protein ARB_00327 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00327 PE=3 SV=2 |
| sp|D4ALG0|LYSM1_ARTBC | 5.76e-30 | 494 | 808 | 45 | 373 | LysM domain-containing protein ARB_05157 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_05157 PE=1 SV=1 |
| sp|P23176|AMYG_ASPKA | 1.17e-19 | 934 | 1038 | 534 | 639 | Glucoamylase I OS=Aspergillus kawachii OX=1069201 GN=gaI PE=1 SV=1 |
| sp|D4AY86|LYSM6_ARTBC | 4.96e-19 | 64 | 437 | 40 | 422 | LysM domain-containing protein ARB_01155/01156 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_01155/01156 PE=3 SV=2 |
| sp|P22832|AMYG_ASPUS | 8.16e-19 | 934 | 1038 | 534 | 639 | Glucoamylase OS=Aspergillus usamii OX=186680 GN=glaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
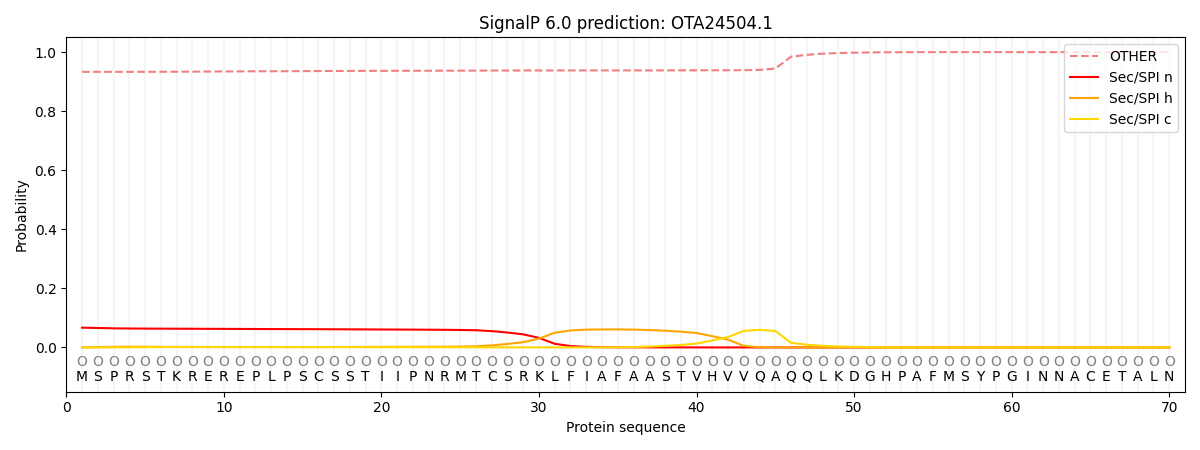
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.936076 | 0.063922 |
