You are browsing environment: FUNGIDB
CAZyme Information: OTA24311.1
You are here: Home > Sequence: OTA24311.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
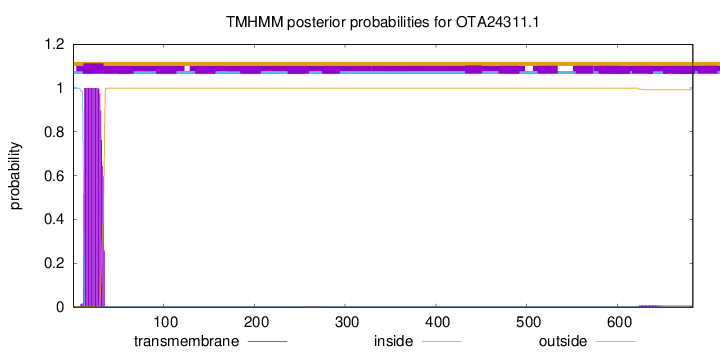
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA24311.1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 180100 | PRK05463 | 1.25e-18 | 508 | 645 | 11 | 133 | putative hydro-lyase. |
| 226786 | YcsI | 5.87e-14 | 508 | 644 | 17 | 139 | Uncharacterized protein YcsI, UPF0317 family [Function unknown]. |
| 367085 | Fringe | 1.21e-08 | 197 | 268 | 72 | 152 | Fringe-like. The drosophila protein fringe (FNG) is a glucosaminyltransferase that controls the response of the Notch receptor to specific ligands. FNG is localized to the Golgi apparatus (not secreted as previously thought). Modification of Notch occurs through glycosylation by FNG. The xenopus homolog, lunatic fringe, has been implicated in a variety of functions. |
| 399927 | DUF1445 | 1.61e-04 | 624 | 644 | 2 | 22 | Protein of unknown function (DUF1445). This family represents a conserved region approximately 150 residues long within a number of hypothetical bacterial and eukaryotic proteins of unknown function. |
| 405052 | PAN_4 | 0.005 | 414 | 457 | 9 | 50 | PAN domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.14e-110 | 54 | 502 | 30 | 473 | |
| 2.81e-109 | 37 | 494 | 30 | 477 | |
| 2.81e-109 | 37 | 494 | 30 | 477 | |
| 2.81e-109 | 37 | 494 | 30 | 477 | |
| 2.81e-109 | 37 | 494 | 30 | 477 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.30e-07 | 502 | 665 | 12 | 155 | Crystal structure of UPF0317 protein PSPTO_5379 from Pseudomonas syringae pv. tomato. NorthEast Structural Genomics target PsR181 [Pseudomonas syringae pv. tomato str. DC3000],2PIF_B Crystal structure of UPF0317 protein PSPTO_5379 from Pseudomonas syringae pv. tomato. NorthEast Structural Genomics target PsR181 [Pseudomonas syringae pv. tomato str. DC3000] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.14e-12 | 505 | 651 | 23 | 154 | Putative hydro-lyase BB2997 OS=Bordetella bronchiseptica (strain ATCC BAA-588 / NCTC 13252 / RB50) OX=257310 GN=BB2997 PE=3 SV=1 |
|
| 1.14e-12 | 505 | 651 | 23 | 154 | Putative hydro-lyase BP1875 OS=Bordetella pertussis (strain Tohama I / ATCC BAA-589 / NCTC 13251) OX=257313 GN=BP1875 PE=3 SV=2 |
|
| 1.14e-12 | 505 | 651 | 23 | 154 | Putative hydro-lyase BPP3031 OS=Bordetella parapertussis (strain 12822 / ATCC BAA-587 / NCTC 13253) OX=257311 GN=BPP3031 PE=3 SV=1 |
|
| 2.34e-12 | 499 | 651 | 4 | 141 | Putative hydro-lyase Bpet2233 OS=Bordetella petrii (strain ATCC BAA-461 / DSM 12804 / CCUG 43448) OX=340100 GN=Bpet2233 PE=3 SV=1 |
|
| 1.36e-11 | 498 | 651 | 2 | 139 | Putative hydro-lyase Cbei_2760 OS=Clostridium beijerinckii (strain ATCC 51743 / NCIMB 8052) OX=290402 GN=Cbei_2760 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
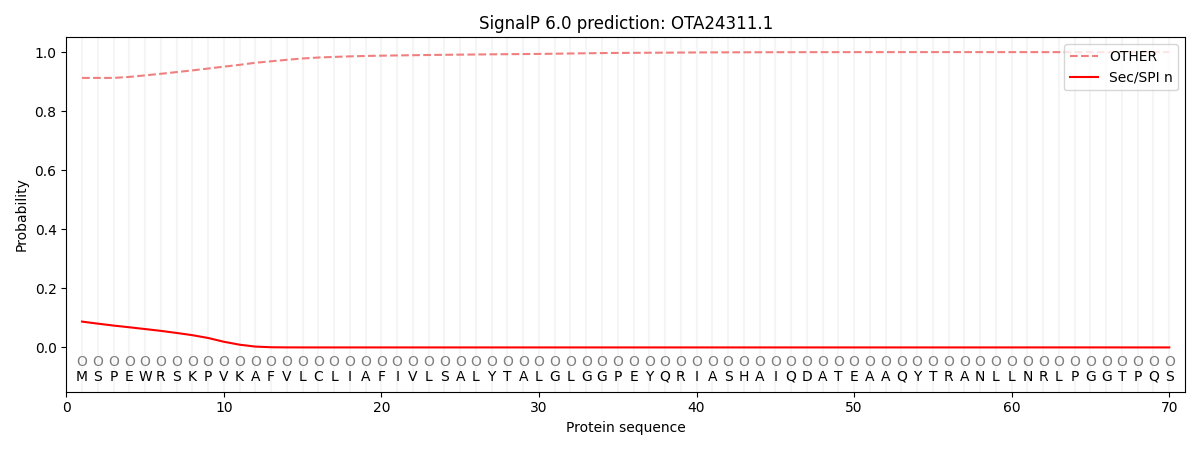
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.912916 | 0.087110 |