You are browsing environment: FUNGIDB
CAZyme Information: OTA23333.1
You are here: Home > Sequence: OTA23333.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA23333.1 | |||||||||||
| CAZy Family | AA7 | |||||||||||
| CAZyme Description | Asparagine-linked glycosylation protein 6 [Source:UniProtKB/TrEMBL;Acc:A0A1Z5SR73] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.258:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT58 | 22 | 218 | 1.3e-78 | 0.5384615384615384 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398745 | ALG3 | 9.29e-114 | 22 | 218 | 1 | 197 | ALG3 protein. The formation of N-glycosidic linkages of glycoproteins involves the ordered assembly of the common Glc3Man9GlcNAc2 core-oligosaccharide on the lipid carrier dolichyl pyrophosphate. Whereas early mannosylation steps occur on the cytoplasmic side of the endoplasmic reticulum with GDP-Man as donor, the final reactions from Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol on the lumenal side use Dol-P-Man. ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. |
| 395416 | Ion_trans | 2.56e-13 | 599 | 826 | 13 | 238 | Ion transport protein. This family contains sodium, potassium and calcium ion channels. This family is 6 transmembrane helices in which the last two helices flank a loop which determines ion selectivity. In some sub-families (e.g. Na channels) the domain is repeated four times, whereas in others (e.g. K channels) the protein forms as a tetramer in the membrane. |
| 400395 | PKD_channel | 4.88e-07 | 571 | 828 | 154 | 423 | Polycystin cation channel. This family contains the cation channel region of PKD1 and PKD2 proteins. |
| 273311 | trp | 8.01e-06 | 480 | 884 | 277 | 668 | transient-receptor-potential calcium channel protein. The Transient Receptor Potential Ca2+ Channel (TRP-CC) Family (TC. 1.A.4)The TRP-CC family has also been called the store-operated calcium channel (SOC) family. The prototypical members include the Drosophila retinal proteinsTRP and TRPL (Montell and Rubin, 1989; Hardie and Minke, 1993). SOC members of the family mediate the entry of extracellular Ca2+ into cells in responseto depletion of intracellular Ca2+ stores (Clapham, 1996) and agonist stimulated production of inositol-1,4,5 trisphosphate (IP3). One member of the TRP-CCfamily, mammalian Htrp3, has been shown to form a tight complex with the IP3 receptor (TC #1.A.3.2.1). This interaction is apparently required for IP3 tostimulate Ca2+ release via Htrp3. The vanilloid receptor subtype 1 (VR1), which is the receptor for capsaicin (the ?hot? ingredient in chili peppers) and servesas a heat-activated ion channel in the pain pathway (Caterina et al., 1997), is also a member of this family. The stretch-inhibitable non-selective cation channel(SIC) is identical to the vanilloid receptor throughout all of its first 700 residues, but it exhibits a different sequence in its last 100 residues. VR1 and SICtransport monovalent cations as well as Ca2+. VR1 is about 10x more permeable to Ca2+ than to monovalent ions. Ca2+ overload probably causes cell deathafter chronic exposure to capsaicin. (McCleskey and Gold, 1999). [Transport and binding proteins, Cations and iron carrying compounds] |
| 177614 | PHA03377 | 0.005 | 961 | 1233 | 361 | 654 | EBNA-3C; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.20e-85 | 1 | 218 | 1 | 218 | |
| 3.25e-82 | 1 | 218 | 1 | 218 | |
| 6.02e-82 | 1 | 218 | 1 | 218 | |
| 6.02e-82 | 1 | 218 | 1 | 218 | |
| 6.02e-82 | 1 | 218 | 1 | 218 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.23e-69 | 1 | 228 | 292 | 521 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Phaeosphaeria nodorum (strain SN15 / ATCC MYA-4574 / FGSC 10173) OX=321614 GN=ALG3 PE=3 SV=2 |
|
| 2.26e-67 | 3 | 228 | 14 | 243 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-3 PE=3 SV=1 |
|
| 2.53e-65 | 1 | 234 | 1 | 229 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=alg3 PE=3 SV=1 |
|
| 8.58e-63 | 1 | 226 | 63 | 286 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=alg3 PE=3 SV=1 |
|
| 2.87e-61 | 1 | 220 | 9 | 228 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=alg3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000014 | 0.000003 |
TMHMM Annotations download full data without filtering help
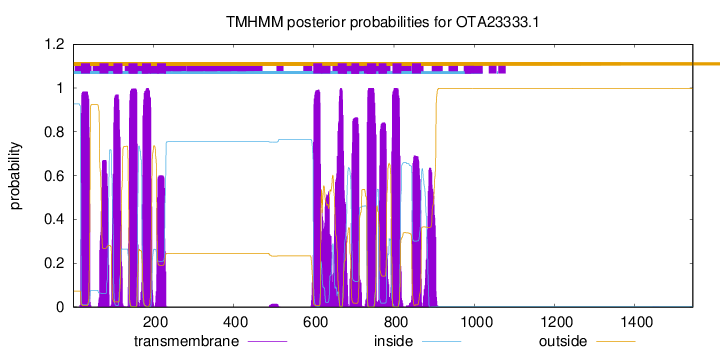
| Start | End |
|---|---|
| 20 | 42 |
| 66 | 88 |
| 101 | 123 |
| 138 | 160 |
| 172 | 194 |
| 209 | 231 |
| 599 | 621 |
| 660 | 682 |
| 694 | 713 |
| 733 | 755 |
| 762 | 781 |
| 796 | 818 |
| 845 | 867 |
