You are browsing environment: FUNGIDB
CAZyme Information: OTA22089.1
You are here: Home > Sequence: OTA22089.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA22089.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8394; End:13480 Strand: - | |||||||||||
Full Sequence Download help
| MYEMELKPGS KRNMLAMTGV ITSTTIKNSI ITVLQDMEYI HGQLFERTVE SGCGAVTCLP | 60 |
| GALTMLRFSA FRRMAKWYFA DKAEQISDLF DWGKCHLGED RWLTHLFMIG ARERYQIQMN | 120 |
| TGAFCKTEAV QTFKSLLKQR RRWFLGFITN EVCMLTDVRL WRRYPVLLVI RLAQNTIRTT | 180 |
| ALLFFIMVIS IVTTSQKISN LPVGFIAVSL GLNWLLMLYF GAKLGRWKIM LYPLMFVLNP | 240 |
| FFNYIYMVYG IFTAGNRTWG GPRADAAIKA DEHTTPAEAI EKAEEEGNEL LLVPETLKPA | 300 |
| SAALVLPSDV ASDKHKTQGL RNKLLASLSV DGIAAYKLTP NVELLCLADA IVTHPAVLKG | 360 |
| VEPARWAKLR TGVCRQRLLS EPSSTLQNLI YDDLDVVEEQ LGKLEGSFKE QAQVEFLLER | 420 |
| AEVDTHHGLD KIARQNLDEA KQRRRFEYAL TGLLGKRTKF QEKDFSQLVV LAKSASNDES | 480 |
| GSTETSDAAP GPVEKEETGP AKLDLNDDTL LEPKESESAS ASERLRYIFQ LASPYGWDLE | 540 |
| AELAARWVQL GGLRSALEIY ERLEMWAEAA LCWAATERED KAKNMIKKQL FHATSGKSDV | 600 |
| EADDDETWEG EERQPPPMDA PRLYCILGDI DQDVSMYEKA WTVSKERYAR AQRSLGRLYI | 660 |
| TKGDMVKAAE AYSKSLRVNQ LNQQSWFALG CALLELAQFE KAVAAFGRCV QIDDMDAEGW | 720 |
| SNLAAALLRC EPDTDDGAKP GAAPADDEDE SVITYREKPD PQKHRQDALK ALKRAAGLKH | 780 |
| DSYRIWENLL IVAASLSPPD YNSALAAQKR IIDIRGKTDG EKCIDKQILE MLASHVITTS | 840 |
| DRYDATQPGL PRIFVKFMDE SVIPLITGSA ELWQLVSKLA LWRNKPSSAL DAEEKSWRAV | 900 |
| TAQPGWETEN EKLWNLVVDA TVRLCEAYES FGPMERTEGL ASGEPVAKDW KFKARSALRG | 960 |
| ILGKAKGTWE GTDGWERLQE TMQGLKA | 987 |
Enzyme Prediction help
| EC | 2.4.1.16:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 14 | 269 | 6.1e-24 | 0.5066413662239089 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133033 | Chitin_synth_C | 1.31e-17 | 2 | 143 | 96 | 237 | C-terminal domain of Chitin Synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin. Chitin synthase, also called UDP-N-acetyl-D-glucosamine:chitin 4-beta-N-acetylglucosaminyltransferase, catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of GlcNAc residues formed by covalent beta-1,4 linkages. Chitin is an important component of the cell wall of fungi and bacteria and it is synthesized on the cytoplasmic surface of the cell membrane by membrane bound chitin synthases. Studies with fungi have revealed that most of them contain more than one chitin synthase gene. At least five subclasses of chitin synthases have been identified. |
| 367353 | Chitin_synth_2 | 3.81e-16 | 19 | 263 | 234 | 493 | Chitin synthase. Members of this family are fungal chitin synthase EC:2.4.1.16 enzymes. They catalyze chitin synthesis as follows: UDP-N-acetyl-D-glucosamine + {(1,4)-(N-acetyl-beta-D-glucosaminyl)}(N) <=> UDP + {(1,4)-(N-acetyl-beta-D-glucosaminyl)}(N+1). |
| 276809 | TPR | 2.65e-13 | 649 | 729 | 1 | 81 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 224136 | BcsA | 2.18e-11 | 31 | 252 | 186 | 397 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 276809 | TPR | 2.20e-10 | 623 | 711 | 10 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBX99330.1|GT2 | 2.52e-167 | 1 | 303 | 265 | 566 |
| CAE7027846.1|GT2 | 7.10e-166 | 1 | 303 | 291 | 592 |
| QRD05648.1|GT2 | 3.91e-164 | 1 | 303 | 292 | 593 |
| QDS70805.1|GT2 | 4.17e-164 | 1 | 301 | 288 | 587 |
| SMR59275.1|GT2 | 4.16e-161 | 2 | 307 | 287 | 591 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P78746|CHSD_ASPFU | 4.00e-150 | 1 | 290 | 234 | 522 | Chitin synthase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=chsD PE=3 SV=1 |
| sp|O36033|YLM1_SCHPO | 1.02e-80 | 320 | 986 | 92 | 817 | TPR repeat-containing protein C19B12.01 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC19B12.01 PE=4 SV=2 |
| sp|P42842|EMW1_YEAST | 8.52e-57 | 502 | 980 | 476 | 899 | Essential for maintenance of the cell wall protein 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=EMW1 PE=1 SV=1 |
| sp|Q8CD92|TTC27_MOUSE | 7.93e-34 | 306 | 699 | 126 | 581 | Tetratricopeptide repeat protein 27 OS=Mus musculus OX=10090 GN=Ttc27 PE=1 SV=2 |
| sp|Q17QZ7|TTC27_BOVIN | 4.23e-33 | 306 | 699 | 126 | 581 | Tetratricopeptide repeat protein 27 OS=Bos taurus OX=9913 GN=TTC27 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
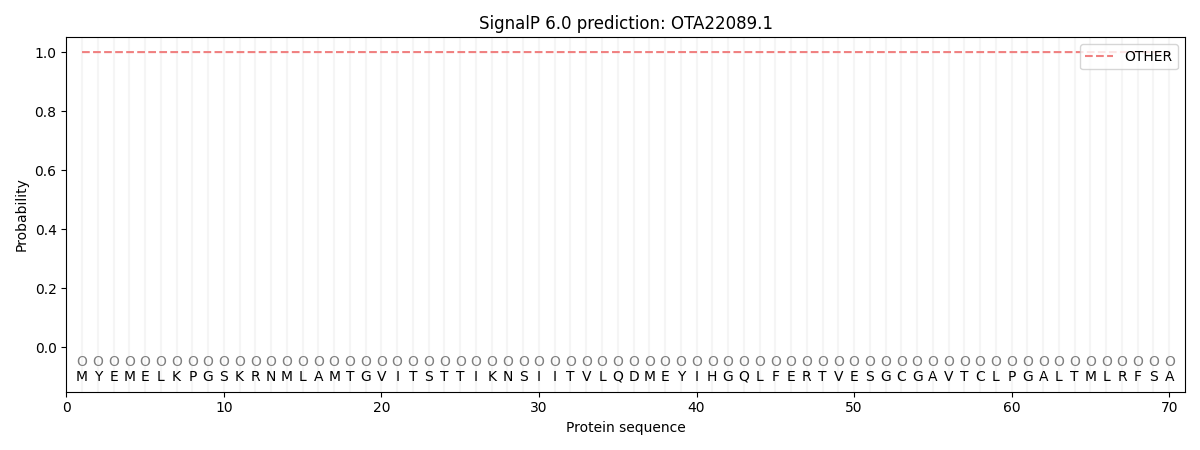
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000052 | 0.000001 |
TMHMM Annotations download full data without filtering help
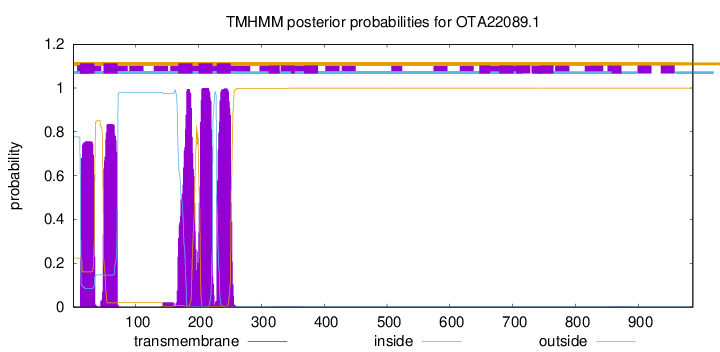
| Start | End |
|---|---|
| 12 | 34 |
| 49 | 71 |
| 168 | 190 |
| 200 | 222 |
| 229 | 251 |
