You are browsing environment: FUNGIDB
CAZyme Information: OTA19153.1
You are here: Home > Sequence: OTA19153.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
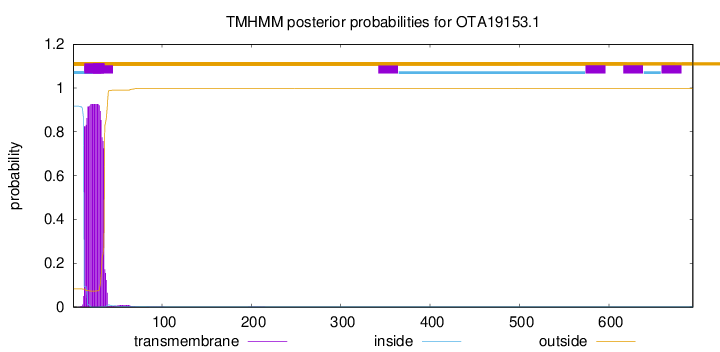
TMHMM annotations
Basic Information help
| Species | Hortaea werneckii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Teratosphaeriaceae; Hortaea; Hortaea werneckii | |||||||||||
| CAZyme ID | OTA19153.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE18 | 257 | 622 | 1.2e-199 | 0.997275204359673 |
| CBM87 | 39 | 256 | 8.5e-108 | 0.9908256880733946 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 200575 | CE4_ClCDA_like | 0.009 | 362 | 424 | 18 | 98 | Catalytic NodB homology domain of Colletotrichum lindemuthianum chitin deacetylase and similar proteins. This family is represented by the chitin deacetylase (endo-chitin de-N-acetylase, ClCDA, EC 3.5.1.41) from Colletotrichum lindemuthianum (also known as Glomerella lindemuthiana), which is a member of the carbohydrate esterase 4 (CE4) superfamily. ClCDA catalyzes the hydrolysis of N-acetamido groups of N-acetyl-D-glucosamine residues in chitin, converting it to chitosan in fungal cell walls. It consists of a single catalytic domain similar to the deformed (alpha/beta)8 barrel fold adopted by other CE4 esterases, which encompasses a mononuclear metalloenzyme employing a conserved His-His-Asp zinc-binding triad closely associated with the conserved catalytic base (aspartic acid) and acid (histidine), to carry out acid/base catalysis. It possesses a highly conserved substrate-binding groove, with subtle alterations that influence substrate specificity and subsite affinity. Unlike its bacterial homologs, ClCDA contains two intramolecular disulfide bonds that may add stability to this secreted protein. The family also includes many uncharacterized deacetylases and hypothetical proteins mainly from eukaryotes, which show high sequence similarity to ClCDA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.38e-309 | 30 | 694 | 42 | 708 | |
| 2.83e-298 | 28 | 694 | 25 | 692 | |
| 4.02e-298 | 28 | 694 | 25 | 692 | |
| 4.02e-298 | 28 | 694 | 25 | 692 | |
| 2.89e-293 | 13 | 692 | 10 | 689 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.85e-273 | 30 | 694 | 22 | 684 | Crystal structure of Agd3 a novel carbohydrate deacetylase [Aspergillus fumigatus Af293] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.147567 | 0.852393 | CS pos: 28-29. Pr: 0.7733 |