You are browsing environment: FUNGIDB
CAZyme Information: ORE09557.1
You are here: Home > Sequence: ORE09557.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus microsporus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus microsporus | |||||||||||
| CAZyme ID | ORE09557.1 | |||||||||||
| CAZy Family | GT31 | |||||||||||
| CAZyme Description | L-ascorbate oxidase [Source:UniProtKB/TrEMBL;Acc:A0A1X0RBZ2] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 40 | 533 | 4.9e-88 | 0.9664804469273743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 274555 | ascorbase | 1.36e-92 | 19 | 547 | 2 | 530 | L-ascorbate oxidase, plant type. Members of this protein family are the copper-containing enzyme L-ascorbate oxidase (EC 1.10.3.3), also called ascorbase. This family is found in flowering plants, and shows greater sequence similarity to a family of laccases (EC 1.10.3.2) from plants than to other known ascorbate oxidases. |
| 177843 | PLN02191 | 1.27e-80 | 4 | 547 | 9 | 553 | L-ascorbate oxidase |
| 215324 | PLN02604 | 8.75e-80 | 4 | 547 | 10 | 553 | oxidoreductase |
| 132431 | ascorbOXfungal | 2.50e-76 | 23 | 533 | 13 | 524 | L-ascorbate oxidase, fungal type. This model describes a family of fungal ascorbate oxidases, within a larger family of multicopper oxidases that also includes plant ascorbate oxidases (TIGR03388), plant laccases and laccase-like proteins (TIGR03389), and related proteins. The member from Acremonium sp. HI-25 is characterized. |
| 274556 | laccase | 3.15e-54 | 16 | 538 | 1 | 518 | laccase, plant. Members of this protein family include the copper-containing enzyme laccase (EC 1.10.3.2), often several from a single plant species, and additional, uncharacterized, closely related plant proteins termed laccase-like multicopper oxidases. This protein family shows considerable sequence similarity to the L-ascorbate oxidase (EC 1.10.3.3) family. Laccases are enzymes of rather broad specificity, and classification of all proteins scoring about the trusted cutoff of this model as laccases may be appropriate. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.29e-168 | 18 | 551 | 25 | 651 | |
| 9.89e-143 | 4 | 545 | 6 | 573 | |
| 3.37e-133 | 21 | 546 | 18 | 571 | |
| 8.14e-108 | 4 | 511 | 6 | 518 | |
| 4.81e-75 | 13 | 547 | 28 | 558 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.10e-64 | 18 | 547 | 3 | 530 | Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1AOZ_B Refined Crystal Structure Of Ascorbate Oxidase At 1.9 Angstroms Resolution [Cucurbita pepo var. melopepo],1ASO_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASO_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASP_A X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASP_B X-ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-forms [Cucurbita pepo var. melopepo],1ASQ_A X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo],1ASQ_B X-Ray Structures And Mechanistic Implications Of Three Functional Derivatives Of Ascorbate Oxidase From Zucchini: Reduced-, Peroxide-, And Azide-Forms [Cucurbita pepo var. melopepo] |
|
| 2.10e-52 | 37 | 542 | 20 | 481 | Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_B Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_C Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_D Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_E Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae],1ZPU_F Crystal Structure of Fet3p, a Multicopper Oxidase that Functions in Iron Import [Saccharomyces cerevisiae] |
|
| 5.99e-40 | 22 | 546 | 7 | 479 | Crystal structure of LacB from Trametes sp. AH28-2 [Trametes sp. AH28-2],3KW7_B Crystal structure of LacB from Trametes sp. AH28-2 [Trametes sp. AH28-2] |
|
| 1.22e-39 | 19 | 546 | 68 | 548 | Structure of the L499M mutant of the laccase from B.aclada [Botrytis aclada] |
|
| 1.66e-39 | 19 | 546 | 68 | 548 | Crystal structure of laccase from Botrytis aclada at 1.67 A resolution [Botrytis aclada],4X4K_A Structure of laccase from Botrytis aclada with full copper content [Botrytis aclada] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.78e-68 | 16 | 547 | 18 | 549 | L-ascorbate oxidase OS=Brassica rapa subsp. pekinensis OX=51351 GN=AO PE=2 SV=1 |
|
| 6.43e-68 | 16 | 547 | 28 | 557 | L-ascorbate oxidase OS=Nicotiana tabacum OX=4097 GN=AAO PE=2 SV=1 |
|
| 3.20e-65 | 16 | 550 | 21 | 553 | L-ascorbate oxidase OS=Arabidopsis thaliana OX=3702 GN=AAO PE=1 SV=1 |
|
| 4.17e-63 | 18 | 547 | 3 | 530 | L-ascorbate oxidase OS=Cucurbita pepo var. melopepo OX=3665 PE=1 SV=1 |
|
| 1.41e-62 | 18 | 547 | 33 | 560 | L-ascorbate oxidase OS=Cucurbita maxima OX=3661 GN=AAO PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
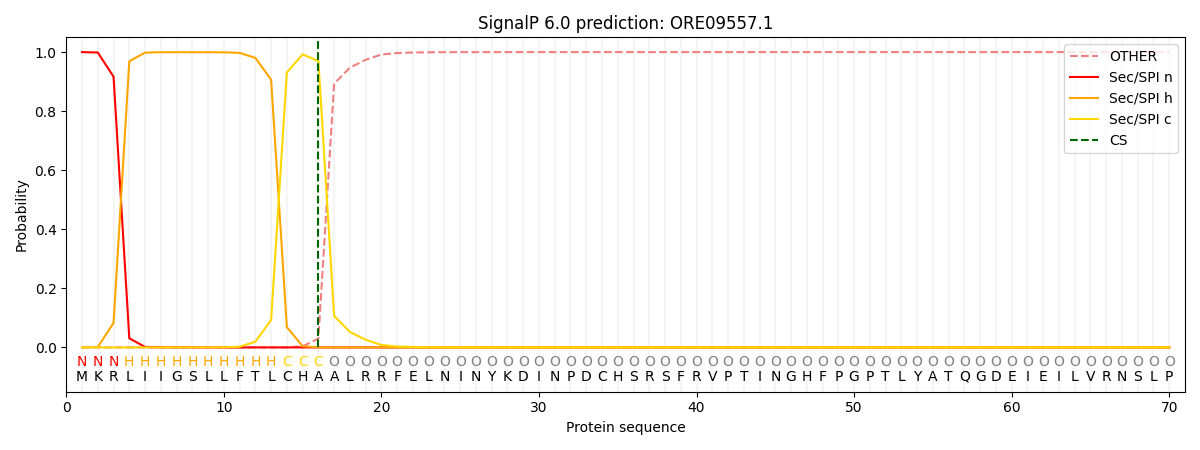
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000716 | 0.999255 | CS pos: 16-17. Pr: 0.9694 |
