You are browsing environment: FUNGIDB
CAZyme Information: ORE09457.1
You are here: Home > Sequence: ORE09457.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus microsporus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus microsporus | |||||||||||
| CAZyme ID | ORE09457.1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | Nucleotid_trans domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1X0RBN6] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT77 | 122 | 327 | 3.8e-24 | 0.9722222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 367480 | Nucleotid_trans | 2.11e-15 | 122 | 326 | 4 | 204 | Nucleotide-diphospho-sugar transferase. Proteins in this family have been been predicted to be nucleotide-diphospho-sugar transferases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.30e-115 | 63 | 346 | 96 | 379 | |
| 7.78e-114 | 54 | 348 | 46 | 344 | |
| 4.23e-90 | 34 | 346 | 49 | 362 | |
| 2.02e-78 | 74 | 337 | 101 | 368 | |
| 4.96e-73 | 92 | 337 | 110 | 356 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.06e-20 | 91 | 358 | 8 | 316 | UDP-galactose:fucoside alpha-3-galactosyltransferase OS=Dictyostelium discoideum OX=44689 GN=agtA PE=1 SV=1 |
|
| 1.31e-09 | 65 | 326 | 242 | 517 | Beta-arabinofuranosyltransferase RAY1 OS=Arabidopsis thaliana OX=3702 GN=RAY1 PE=2 SV=1 |
|
| 1.19e-08 | 82 | 343 | 81 | 354 | UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1 OS=Arabidopsis thaliana OX=3702 GN=RGXT1 PE=1 SV=1 |
|
| 9.60e-08 | 88 | 343 | 77 | 344 | UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 3 OS=Arabidopsis thaliana OX=3702 GN=RGXT3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
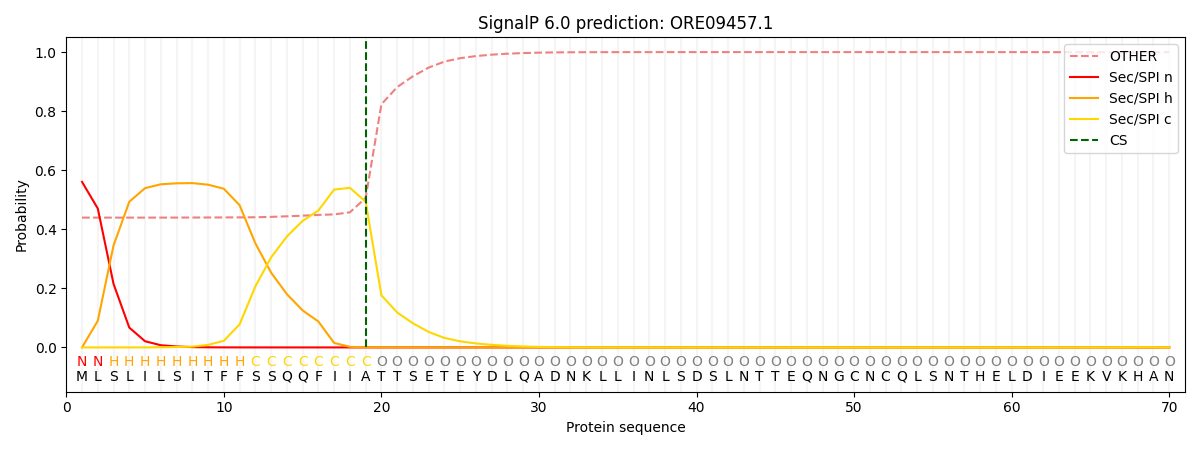
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.455414 | 0.544592 | CS pos: 19-20. Pr: 0.4935 |
