You are browsing environment: FUNGIDB
CAZyme Information: ORE09356.1
You are here: Home > Sequence: ORE09356.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
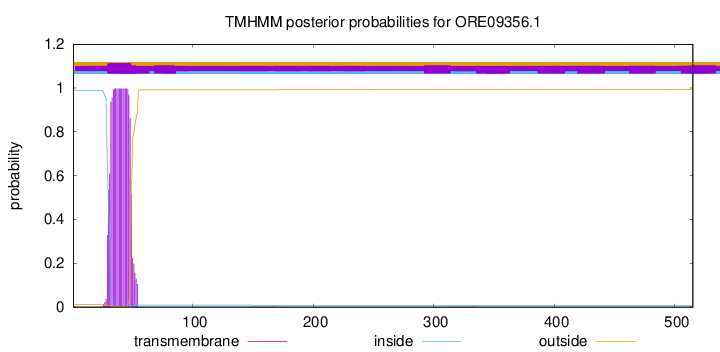
TMHMM annotations
Basic Information help
| Species | Rhizopus microsporus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus microsporus | |||||||||||
| CAZyme ID | ORE09356.1 | |||||||||||
| CAZy Family | GT24 | |||||||||||
| CAZyme Description | Glycosyltransferase-like protein LARGE1 [Source:UniProtKB/TrEMBL;Acc:A0A1X0RBI0] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:23 | 2.4.2.-:23 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT49 | 182 | 487 | 2.1e-51 | 0.8694362017804155 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404735 | Glyco_transf_49 | 3.91e-39 | 198 | 483 | 1 | 326 | Glycosyl-transferase for dystroglycan. This glycosyl-transferase brings about the glycosylation of the alpha-dystroglycan subunit. Dystroglycan is an integral member of the skeletal muscular dystrophin glycoprotein complex, which links dystrophin to proteins in the extracellular matrix. |
| 133035 | GT_2_like_e | 0.004 | 281 | 360 | 77 | 162 | Subfamily of Glycosyltransferase Family GT2 of unknown function. GT-2 includes diverse families of glycosyltransferases with a common GT-A type structural fold, which has two tightly associated beta/alpha/beta domains that tend to form a continuous central sheet of at least eight beta-strands. These are enzymes that catalyze the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. Glycosyltransferases have been classified into more than 90 distinct sequence based families. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.77e-145 | 63 | 512 | 85 | 528 | |
| 3.24e-65 | 95 | 495 | 118 | 564 | |
| 1.05e-62 | 198 | 495 | 207 | 554 | |
| 9.67e-30 | 183 | 450 | 42 | 293 | |
| 4.79e-29 | 183 | 450 | 304 | 555 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.21e-29 | 183 | 450 | 458 | 709 | Xylosyl- and glucuronyltransferase LARGE1 OS=Mus musculus OX=10090 GN=Large1 PE=1 SV=1 |
|
| 8.34e-29 | 183 | 484 | 459 | 744 | Xylosyl- and glucuronyltransferase LARGE1 OS=Danio rerio OX=7955 GN=large1 PE=2 SV=1 |
|
| 1.29e-28 | 198 | 450 | 402 | 639 | Xylosyl- and glucuronyltransferase LARGE2 OS=Mus musculus OX=10090 GN=Large2 PE=1 SV=1 |
|
| 3.47e-28 | 198 | 450 | 456 | 692 | Xylosyl- and glucuronyltransferase LARGE2s OS=Gallus gallus OX=9031 GN=LARGE2 PE=2 SV=1 |
|
| 4.18e-28 | 198 | 450 | 401 | 639 | Xylosyl- and glucuronyltransferase LARGE2 OS=Rattus norvegicus OX=10116 GN=Large2 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000017 | 0.000004 |