You are browsing environment: FUNGIDB
CAZyme Information: ORE06703.1
You are here: Home > Sequence: ORE06703.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus microsporus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus microsporus | |||||||||||
| CAZyme ID | ORE06703.1 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | UDP-Glycosyltransferase/glycogen phosphorylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT1 | 260 | 458 | 4.3e-27 | 0.3717277486910995 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340817 | GT1_Gtf-like | 6.89e-52 | 30 | 477 | 2 | 403 | UDP-glycosyltransferases and similar proteins. This family includes the Gtfs, a group of homologous glycosyltransferases involved in the final stages of the biosynthesis of antibiotics vancomycin and related chloroeremomycin. Gtfs transfer sugar moieties from an activated NDP-sugar donor to the oxidatively cross-linked heptapeptide core of vancomycin group antibiotics. The core structure is important for the bioactivity of the antibiotics. |
| 278624 | UDPGT | 1.42e-30 | 41 | 519 | 11 | 478 | UDP-glucoronosyl and UDP-glucosyl transferase. |
| 224732 | YjiC | 4.09e-26 | 28 | 477 | 1 | 397 | UDP:flavonoid glycosyltransferase YjiC, YdhE family [Carbohydrate transport and metabolism]. |
| 223071 | egt | 1.03e-21 | 243 | 459 | 250 | 445 | ecdysteroid UDP-glucosyltransferase; Provisional |
| 273616 | MGT | 3.48e-21 | 36 | 476 | 2 | 387 | glycosyltransferase, MGT family. This model describes the MGT (macroside glycosyltransferase) subfamily of the UDP-glucuronosyltransferase family. Members include a number of glucosyl transferases for macrolide antibiotic inactivation, but also include transferases of glucose-related sugars for macrolide antibiotic production. [Cellular processes, Toxin production and resistance] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.85e-212 | 1 | 519 | 1 | 524 | |
| 1.15e-105 | 21 | 516 | 25 | 519 | |
| 1.51e-95 | 20 | 519 | 34 | 531 | |
| 9.71e-92 | 20 | 516 | 28 | 522 | |
| 8.02e-86 | 8 | 508 | 18 | 515 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.54e-15 | 235 | 422 | 212 | 393 | Structure of UGT78G1 complexed with myricetin and UDP [Medicago truncatula],3HBJ_A Structure of UGT78G1 complexed with UDP [Medicago truncatula] |
|
| 2.58e-14 | 252 | 441 | 240 | 415 | Crystal Structure of Medicago truncatula UGT71G1 [Medicago truncatula],2ACV_B Crystal Structure of Medicago truncatula UGT71G1 [Medicago truncatula] |
|
| 2.60e-14 | 252 | 441 | 240 | 415 | Crystal Structure of Medicago truncatula UGT71G1 complexed with UDP-glucose [Medicago truncatula],2ACW_B Crystal Structure of Medicago truncatula UGT71G1 complexed with UDP-glucose [Medicago truncatula] |
|
| 1.84e-13 | 236 | 448 | 211 | 416 | Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification [Vitis vinifera],2C1Z_A Structure and activity of a flavonoid 3-O glucosyltransferase reveals the basis for plant natural product modification [Vitis vinifera],2C9Z_A Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification [Vitis vinifera] |
|
| 2.01e-13 | 226 | 452 | 198 | 446 | Crystal structure of TcCGT1 in complex with UDP [Trollius chinensis],6JTD_B Crystal structure of TcCGT1 in complex with UDP [Trollius chinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.53e-20 | 41 | 511 | 7 | 470 | UDP-glucuronosyltransferase 2C1 (Fragment) OS=Oryctolagus cuniculus OX=9986 GN=UGT2C1 PE=2 SV=1 |
|
| 3.31e-19 | 224 | 511 | 230 | 492 | UDP-glucuronosyltransferase 3A1 OS=Mus musculus OX=10090 GN=Ugt3a1 PE=1 SV=1 |
|
| 5.88e-19 | 128 | 511 | 131 | 492 | UDP-glucuronosyltransferase 3A1 OS=Homo sapiens OX=9606 GN=UGT3A1 PE=2 SV=1 |
|
| 1.89e-18 | 28 | 513 | 13 | 499 | UDP-glucuronosyltransferase 2B33 OS=Macaca mulatta OX=9544 GN=UGT2B33 PE=1 SV=1 |
|
| 1.94e-18 | 135 | 447 | 123 | 433 | Anthocyanidin 3-O-glucosyltransferase OS=Zea mays OX=4577 GN=BZ1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
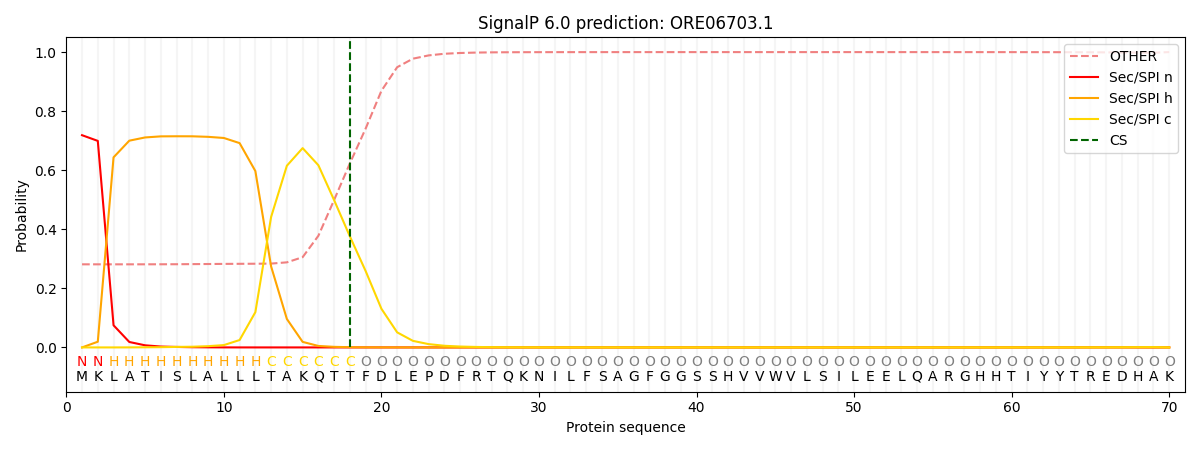
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.300439 | 0.699558 | CS pos: 18-19. Pr: 0.3758 |
