You are browsing environment: FUNGIDB
CAZyme Information: ORE05826.1
You are here: Home > Sequence: ORE05826.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Rhizopus microsporus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Mucoromycota; Mucoromycetes; ; Rhizopodaceae; Rhizopus; Rhizopus microsporus | |||||||||||
| CAZyme ID | ORE05826.1 | |||||||||||
| CAZy Family | GH45 | |||||||||||
| CAZyme Description | chitinase 2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.14:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 30 | 283 | 6.1e-28 | 0.6993243243243243 |
| CBM19 | 353 | 396 | 8.8e-23 | 0.9555555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 119356 | GH18_hevamine_XipI_class_III | 2.02e-119 | 29 | 309 | 1 | 280 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 395573 | Glyco_hydro_18 | 6.59e-26 | 30 | 305 | 1 | 277 | Glycosyl hydrolases family 18. |
| 112252 | CBM_19 | 2.10e-21 | 352 | 399 | 14 | 61 | Carbohydrate binding domain (family 19). |
| 119350 | GH18_chitinase_D-like | 6.60e-18 | 93 | 299 | 50 | 304 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| 119363 | GH18_CTS3_chitinase | 1.07e-17 | 107 | 293 | 69 | 252 | GH18 domain of CTS3 (chitinase 3), an uncharacterized protein from the human fungal pathogen Coccidioides posadasii. CTS3 has a chitinase-like glycosyl hydrolase family 18 (GH18) domain; and has homologs in bacteria as well as fungi. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 541 | 1 | 542 | |
| 0.0 | 1 | 541 | 1 | 540 | |
| 5.07e-195 | 1 | 540 | 1 | 492 | |
| 9.96e-144 | 21 | 540 | 20 | 568 | |
| 6.60e-99 | 9 | 309 | 5 | 316 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.61e-81 | 26 | 300 | 3 | 272 | ScCTS1_apo crystal structure [Saccharomyces cerevisiae],2UY3_A ScCTS1_8-chlorotheophylline crystal structure [Saccharomyces cerevisiae],2UY4_A ScCTS1_acetazolamide crystal structure [Saccharomyces cerevisiae],2UY5_A ScCTS1_kinetin crystal structure [Saccharomyces cerevisiae],4TXE_A ScCTS1 in complex with compound 5 [Saccharomyces cerevisiae] |
|
| 1.36e-51 | 31 | 310 | 3 | 273 | CRYSTAL STRUCTURES OF HEVAMINE, A PLANT DEFENCE PROTEIN WITH CHITINASE AND LYSOZYME ACTIVITY, AND ITS COMPLEX WITH AN INHIBITOR [Hevea brasiliensis],1LLO_A Chain A, HEVAMINE [Hevea brasiliensis],2HVM_A Hevamine A At 1.8 Angstrom Resolution [Hevea brasiliensis] |
|
| 1.86e-50 | 31 | 293 | 3 | 254 | cDNA cloning and 1.75A crystal structure determination of PPL2, a novel chimerolectin from Parkia platycephala seeds exhibiting endochitinolytic activity [Parkia platycephala] |
|
| 7.45e-50 | 31 | 310 | 3 | 273 | Chain A, Hevamine A [Hevea brasiliensis] |
|
| 1.45e-49 | 31 | 310 | 3 | 273 | Chain A, Hevamine A [Hevea brasiliensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 1 | 541 | 1 | 540 | Chitinase 1 OS=Rhizopus oligosporus OX=4847 GN=CHI1 PE=1 SV=1 |
|
| 0.0 | 1 | 541 | 1 | 542 | Chitinase 2 OS=Rhizopus oligosporus OX=4847 GN=CHI2 PE=1 SV=1 |
|
| 9.01e-196 | 1 | 540 | 1 | 492 | Chitinase 1 OS=Rhizopus niveus OX=4844 GN=CHI1 PE=3 SV=1 |
|
| 2.51e-82 | 6 | 311 | 3 | 310 | Chitinase 3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=CHT3 PE=1 SV=2 |
|
| 9.18e-78 | 30 | 316 | 19 | 293 | Chitinase 1 OS=Candida albicans OX=5476 GN=CHT1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
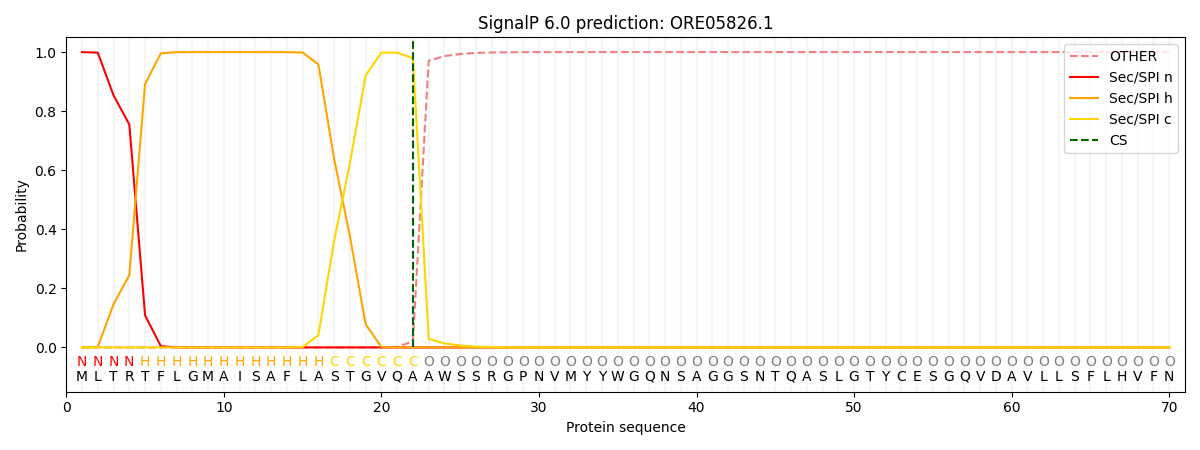
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000223 | 0.999766 | CS pos: 22-23. Pr: 0.9794 |
