You are browsing environment: FUNGIDB
CAZyme Information: OON07390.1
You are here: Home > Sequence: OON07390.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Batrachochytrium salamandrivorans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; NA; Batrachochytrium; Batrachochytrium salamandrivorans | |||||||||||
| CAZyme ID | OON07390.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH46 | 37 | 227 | 9.4e-34 | 0.8423423423423423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 381600 | chitosanase_GH46 | 1.47e-33 | 38 | 223 | 3 | 186 | chitosanase belonging to the glycosyl hydrolase 46 family. This family is composed of the chitosanase enzymes which hydrolyzes chitosan, a biopolymer of beta (1,4)-linked-D-glucosamine (GlcN) residues produced by partial or full deacetylation of chitin. Chitosanases play a role in defense against pathogens such as fungi and are found in microorganisms, fungi, viruses, and plants. Microbial chitosanases can be divided into 3 subclasses based on the specificity of the cleavage positions for partial acetylated chitosan. Subclass I chitosanases such as N174 can split GlcN-GlcN and GlcNAc-GlcN linkages, whereas subclass II chitosanases such as Bacillus sp. no. 7-M can cleave only GlcN-GlcN linkages. Subclass III chitosanases such as MH-K1 chitosanase are the most versatile and can split both GlcN-GlcN and GlcN-GlcNAc linkages. |
| 396099 | Glyco_hydro_46 | 2.31e-21 | 44 | 227 | 1 | 179 | Glycosyl hydrolase family 46. This family are chitosanase enzymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.33e-21 | 44 | 228 | 57 | 242 | |
| 1.66e-19 | 44 | 229 | 42 | 223 | |
| 1.83e-19 | 44 | 229 | 48 | 229 | |
| 1.98e-19 | 44 | 228 | 16 | 195 | |
| 2.71e-19 | 44 | 229 | 13 | 194 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.53e-20 | 44 | 229 | 13 | 194 | Chain A, Chitosanase [Bacillus subtilis subsp. subtilis str. 168] |
|
| 1.84e-19 | 44 | 229 | 13 | 194 | Chain A, Chitosanase [Bacillus subtilis subsp. subtilis str. 168] |
|
| 1.16e-12 | 38 | 223 | 21 | 201 | Chitosanase complex structure [Pseudomonas sp. LL2(2010)],4OLT_B Chitosanase complex structure [Pseudomonas sp. LL2(2010)] |
|
| 1.08e-11 | 41 | 223 | 26 | 203 | Abundantly secreted chitosanase from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4ILY_B Abundantly secreted chitosanase from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
|
| 1.45e-11 | 38 | 223 | 21 | 201 | co-crystal structure of chitosanase OU01 with substrate [Pseudomonas sp. A-01],4QWP_B co-crystal structure of chitosanase OU01 with substrate [Pseudomonas sp. A-01] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.33e-19 | 44 | 229 | 48 | 229 | Chitosanase OS=Bacillus subtilis (strain 168) OX=224308 GN=csn PE=1 SV=1 |
|
| 9.40e-07 | 41 | 229 | 53 | 236 | Chitosanase OS=Streptomyces sp. (strain N174) OX=69019 GN=csn PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
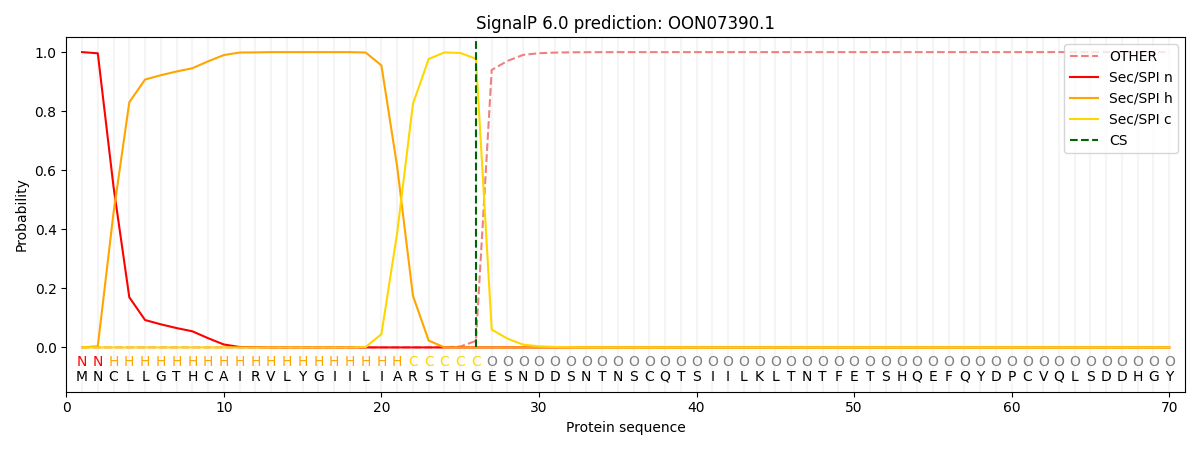
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000214 | 0.999734 | CS pos: 26-27. Pr: 0.9777 |
