You are browsing environment: FUNGIDB
CAZyme Information: OON01126.1
You are here: Home > Sequence: OON01126.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
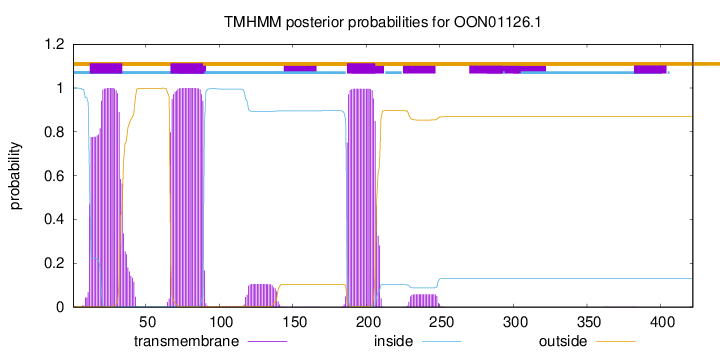
TMHMM annotations
Basic Information help
| Species | Batrachochytrium salamandrivorans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Chytridiomycota; Chytridiomycetes; ; NA; Batrachochytrium; Batrachochytrium salamandrivorans | |||||||||||
| CAZyme ID | OON01126.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | Mannosyltransferase [Source:UniProtKB/TrEMBL;Acc:A0A1S8VGH5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 5 | 249 | 1.7e-46 | 0.5424164524421594 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215437 | PLN02816 | 1.94e-40 | 28 | 285 | 253 | 483 | mannosyltransferase |
| 281842 | Glyco_transf_22 | 4.59e-26 | 14 | 249 | 206 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.40e-50 | 29 | 293 | 269 | 521 | |
| 3.46e-49 | 1 | 415 | 130 | 435 | |
| 9.87e-47 | 4 | 290 | 234 | 487 | |
| 5.68e-46 | 28 | 285 | 97 | 327 | |
| 5.68e-46 | 28 | 285 | 97 | 327 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.87e-45 | 28 | 285 | 253 | 483 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
|
| 1.18e-40 | 29 | 414 | 291 | 628 | GPI mannosyltransferase 3 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=gpi10 PE=3 SV=1 |
|
| 3.17e-40 | 29 | 298 | 272 | 496 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
|
| 1.24e-39 | 44 | 289 | 221 | 434 | GPI mannosyltransferase 3 OS=Drosophila melanogaster OX=7227 GN=PIG-B PE=2 SV=2 |
|
| 4.25e-39 | 29 | 299 | 260 | 479 | GPI mannosyltransferase 3 OS=Xenopus laevis OX=8355 GN=pigb PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
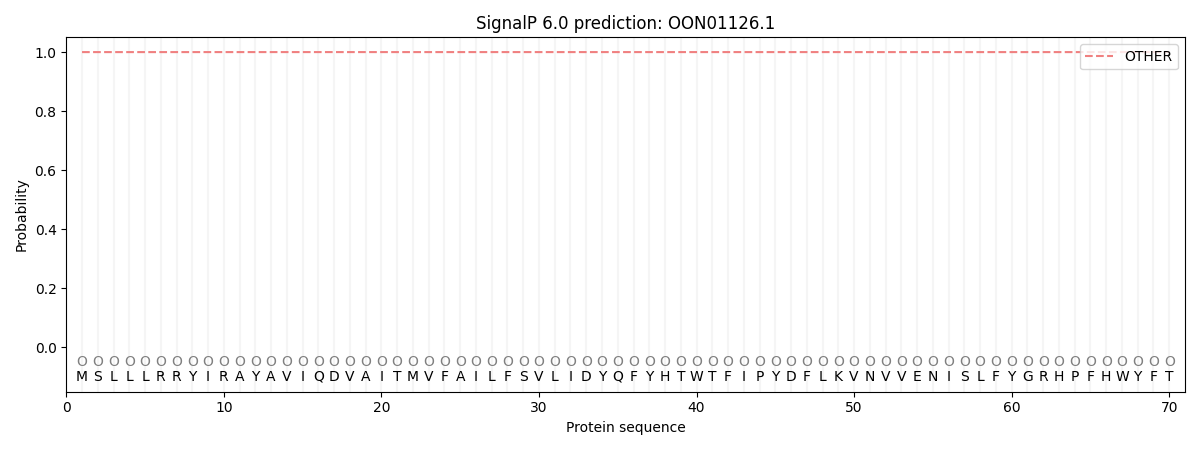
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000031 | 0.000001 |