You are browsing environment: FUNGIDB
CAZyme Information: ONH65604.1
You are here: Home > Sequence: ONH65604.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cyberlindnera fabianii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Phaffomycetaceae; Cyberlindnera; Cyberlindnera fabianii | |||||||||||
| CAZyme ID | ONH65604.1 | |||||||||||
| CAZy Family | GH132 | |||||||||||
| CAZyme Description | putative NAD(P)H dehydrogenase (quinone) FQR1-like 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA6 | 3 | 188 | 3.8e-43 | 0.9897435897435898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223728 | WrbA | 4.47e-27 | 1 | 188 | 3 | 203 | Multimeric flavodoxin WrbA [Energy production and conversion]. |
| 179647 | PRK03767 | 2.39e-21 | 1 | 188 | 1 | 196 | NAD(P)H:quinone oxidoreductase; Provisional |
| 397438 | FMN_red | 8.06e-10 | 2 | 127 | 1 | 140 | NADPH-dependent FMN reductase. |
| 223503 | NorV | 4.10e-07 | 3 | 101 | 248 | 340 | Flavorubredoxin [Energy production and conversion]. |
| 223788 | FldA | 1.37e-06 | 1 | 106 | 1 | 92 | Flavodoxin [Energy production and conversion]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.68e-39 | 2 | 190 | 16 | 200 | |
| 1.68e-39 | 2 | 190 | 16 | 200 | |
| 1.68e-39 | 2 | 190 | 16 | 200 | |
| 1.68e-39 | 2 | 190 | 16 | 200 | |
| 1.16e-26 | 1 | 191 | 2 | 200 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.98e-30 | 6 | 191 | 11 | 188 | The crystal structure of the flavodoxin, WrbA-like protein from Agrobacterium tumefaciens [Agrobacterium fabrum str. C58] |
|
| 1.13e-16 | 1 | 187 | 1 | 193 | Crystal structure of E. coli WrbA in complex with FMN [Escherichia coli],2R96_C Crystal structure of E. coli WrbA in complex with FMN [Escherichia coli],2R97_A Crystal structure of E. coli WrbA in complex with FMN [Escherichia coli],2R97_C Crystal structure of E. coli WrbA in complex with FMN [Escherichia coli],2RG1_A Crystal structure of E. coli WrbA apoprotein [Escherichia coli],2RG1_B Crystal structure of E. coli WrbA apoprotein [Escherichia coli],3B6I_A WrbA from Escherichia coli, native structure [Escherichia coli K-12],3B6I_B WrbA from Escherichia coli, native structure [Escherichia coli K-12],3B6J_A WrbA from Escherichia coli, NADH complex [Escherichia coli K-12],3B6J_B WrbA from Escherichia coli, NADH complex [Escherichia coli K-12],3B6K_A WrbA from Escherichia coli, Benzoquinone complex [Escherichia coli K-12],3B6K_B WrbA from Escherichia coli, Benzoquinone complex [Escherichia coli K-12],3B6M_A WrbA from Escherichia coli, second crystal form [Escherichia coli K-12],3B6M_B WrbA from Escherichia coli, second crystal form [Escherichia coli K-12] |
|
| 4.19e-16 | 3 | 187 | 2 | 192 | High resolution structure of E.coli WrbA with FMN [Escherichia coli APEC O1],4DY4_C High resolution structure of E.coli WrbA with FMN [Escherichia coli APEC O1] |
|
| 8.15e-16 | 3 | 187 | 2 | 192 | X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution [Escherichia coli],3ZHO_B X-ray structure of E.coli Wrba in complex with FMN at 1.2 A resolution [Escherichia coli],4YQE_A Crystal structure of E. coli WrbA in complex with benzoquinone [Escherichia coli K-12],4YQE_B Crystal structure of E. coli WrbA in complex with benzoquinone [Escherichia coli K-12],5F12_A WrbA in complex with FMN under crystallization conditions of WrbA-FMN-BQ structure (4YQE) [Escherichia coli K-12],5F12_B WrbA in complex with FMN under crystallization conditions of WrbA-FMN-BQ structure (4YQE) [Escherichia coli K-12] |
|
| 1.68e-11 | 1 | 186 | 4 | 196 | Structure of B. abortus WrbA-related protein A (WrpA) [Brucella abortus 2308],5F4B_B Structure of B. abortus WrbA-related protein A (WrpA) [Brucella abortus 2308] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.65e-24 | 1 | 188 | 1 | 180 | Flavodoxin FldP OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=fldP PE=2 SV=1 |
|
| 1.92e-21 | 3 | 188 | 4 | 197 | Probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 OS=Arabidopsis thaliana OX=3702 GN=At4g27270 PE=1 SV=1 |
|
| 3.69e-21 | 3 | 188 | 4 | 197 | NAD(P)H dehydrogenase (quinone) FQR1 OS=Arabidopsis thaliana OX=3702 GN=FQR1 PE=1 SV=1 |
|
| 6.79e-21 | 1 | 188 | 1 | 196 | NAD(P)H dehydrogenase (quinone) OS=Hydrogenovibrio crunogenus (strain DSM 25203 / XCL-2) OX=317025 GN=Tcr_1010 PE=3 SV=1 |
|
| 2.86e-18 | 1 | 187 | 1 | 195 | NAD(P)H dehydrogenase (quinone) OS=Burkholderia lata (strain ATCC 17760 / DSM 23089 / LMG 22485 / NCIMB 9086 / R18194 / 383) OX=482957 GN=Bcep18194_C7634 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
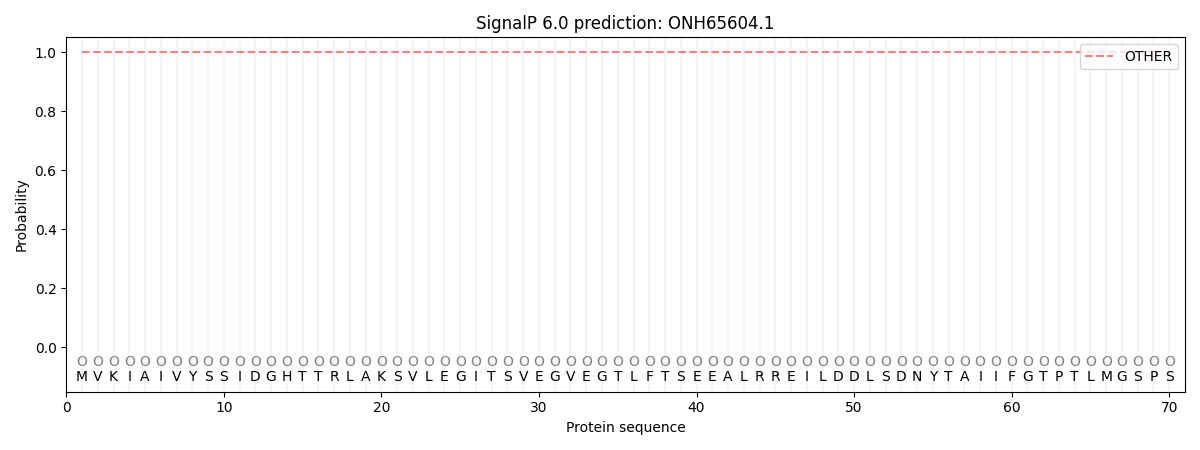
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000063 | 0.000001 |
