You are browsing environment: FUNGIDB
OJD27966.1
Basic Information
help
Species
Blastomyces percursus
Lineage
Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Blastomyces; Blastomyces percursus
CAZyme ID
OJD27966.1
CAZy Family
GT71
CAZyme Description
LysM domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A1J9QGK6]
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
472
50678.45
7.8470
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_BpercursusEI222
10384
N/A
99
10285
Gene Location
No EC number prediction in OJD27966.1.
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
396179
LysM
0.006
229
272
1
43
LysM domain. The LysM (lysin motif) domain is about 40 residues long. It is found in a variety of enzymes involved in bacterial cell wall degradation. This domain may have a general peptidoglycan binding function. The structure of this domain is known.
more
197609
LysM
0.007
229
265
2
38
Lysin motif.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
5.57e-199
1
472
1
495
1.20e-56
13
472
7
438
3.84e-45
61
472
82
463
4.99e-44
14
471
26
470
4.14e-43
14
471
2
457
OJD27966.1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
3.10e-64
66
472
86
463
Uncharacterized protein AN12072 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=AN12072 PE=4 SV=1
more
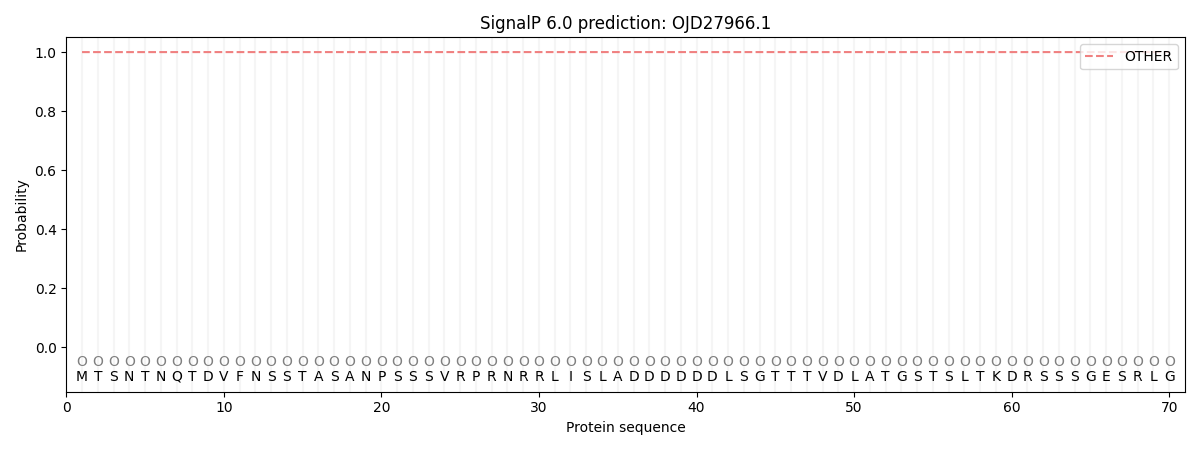
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000043
0.000001
There is no transmembrane helices in OJD27966.1.