You are browsing environment: FUNGIDB
CAZyme Information: OJD26471.1
You are here: Home > Sequence: OJD26471.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
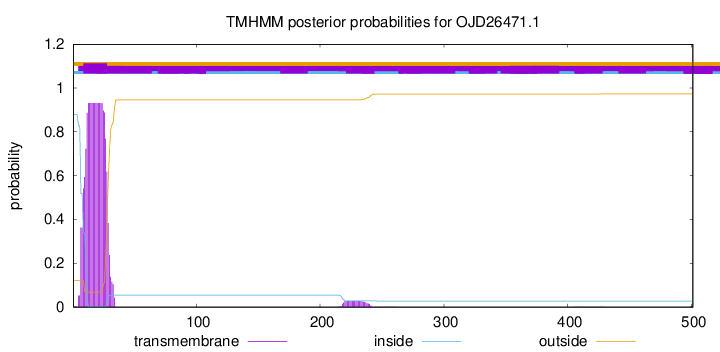
TMHMM annotations
Basic Information help
| Species | Blastomyces percursus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Blastomyces; Blastomyces percursus | |||||||||||
| CAZyme ID | OJD26471.1 | |||||||||||
| CAZy Family | GT21 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT61 | 309 | 426 | 4.7e-26 | 0.9411764705882353 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398330 | DUF563 | 3.31e-25 | 249 | 451 | 15 | 203 | Protein of unknown function (DUF563). Family of uncharacterized proteins. |
| 226845 | COG4421 | 1.20e-05 | 355 | 484 | 229 | 353 | Capsular polysaccharide biosynthesis protein [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.82e-294 | 1 | 501 | 1 | 501 | |
| 9.72e-294 | 1 | 501 | 1 | 501 | |
| 7.83e-287 | 1 | 501 | 1 | 495 | |
| 1.40e-129 | 37 | 500 | 42 | 491 | |
| 4.34e-128 | 40 | 500 | 40 | 490 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.28e-06 | 268 | 431 | 158 | 325 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9J_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_A Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_D Chain D, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9K_E Chain E, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9L_A Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus],7E9L_B Chain B, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Bos taurus] |
|
| 1.68e-06 | 268 | 431 | 148 | 315 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 2.22e-06 | 268 | 431 | 152 | 319 | Chain A, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 3.88e-06 | 268 | 431 | 152 | 319 | Chain D, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
|
| 5.09e-06 | 268 | 431 | 148 | 315 | Chain C, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.50e-17 | 206 | 450 | 219 | 451 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Drosophila melanogaster OX=7227 GN=Eogt PE=1 SV=1 |
|
| 1.37e-13 | 215 | 452 | 241 | 467 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Mus musculus OX=10090 GN=Eogt PE=1 SV=1 |
|
| 1.81e-13 | 215 | 452 | 239 | 465 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Xenopus laevis OX=8355 GN=eogt PE=2 SV=1 |
|
| 1.82e-13 | 271 | 452 | 286 | 467 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Canis lupus familiaris OX=9615 GN=EOGT PE=2 SV=1 |
|
| 1.85e-13 | 261 | 452 | 280 | 475 | EGF domain-specific O-linked N-acetylglucosamine transferase OS=Gallus gallus OX=9031 GN=EOGT PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.986317 | 0.013707 |