You are browsing environment: FUNGIDB
CAZyme Information: OJD16685.1
You are here: Home > Sequence: OJD16685.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
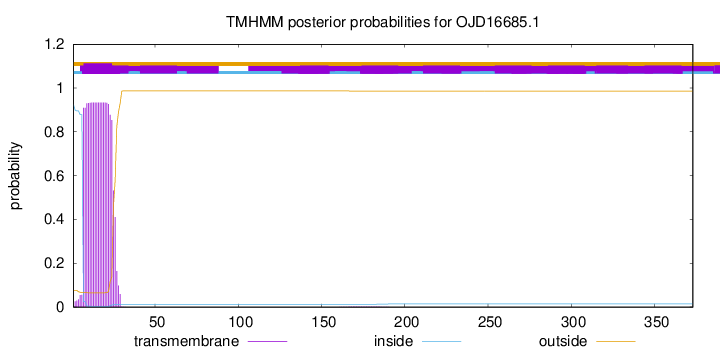
TMHMM annotations
Basic Information help
| Species | Emergomyces pasteurianus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Emergomyces; Emergomyces pasteurianus | |||||||||||
| CAZyme ID | OJD16685.1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.257:12 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226297 | OCH1 | 1.11e-46 | 83 | 361 | 63 | 338 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 398274 | Gly_transf_sug | 1.89e-18 | 129 | 208 | 10 | 90 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 338078 | AvrE | 3.07e-04 | 28 | 106 | 81 | 153 | Pathogenicity factor. This family is secreted by gram-negative Gammaproteobacteria such as Pseudomonas syringae of tomato and the fire blight plant pathogen Erwinia amylovora, amongst others. It is an essential pathogenicity factor of approximately 198 kDa. Its injection into the host-plant is dependent upon the bacterial type III or Hrp secretion system. The family is long and carries a number of predicted functional regions, including in Erwinia stewartii, an ERMS or endoplasmic reticulum membrane retention signal at both the C- and the N-termini, a leucine-zipper motif from residues 539-560, and a nuclear localization signal at 1358-1361. this conserved AvrE-family of effectors is among the few that are required for full virulence of many phytopathogenic pseudomonads, erwinias and pantoeas. A double beta-propeller structure is found towards the N-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.61e-174 | 1 | 370 | 1 | 366 | |
| 5.40e-173 | 1 | 370 | 1 | 366 | |
| 5.48e-152 | 1 | 370 | 1 | 353 | |
| 1.26e-151 | 63 | 367 | 17 | 321 | |
| 6.45e-147 | 1 | 370 | 1 | 356 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.90e-84 | 86 | 370 | 83 | 376 | Initiation-specific alpha-1,6-mannosyltransferase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=OCH1 PE=3 SV=1 |
|
| 1.68e-69 | 79 | 364 | 103 | 382 | Putative glycosyltransferase HOC1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=HOC1 PE=1 SV=3 |
|
| 1.03e-67 | 92 | 370 | 132 | 394 | Initiation-specific alpha-1,6-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=och1 PE=1 SV=2 |
|
| 1.18e-62 | 86 | 370 | 80 | 470 | Initiation-specific alpha-1,6-mannosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=OCH1 PE=1 SV=1 |
|
| 2.83e-06 | 103 | 219 | 58 | 177 | Inositol phosphoceramide mannosyltransferase 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC17G8.11c PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
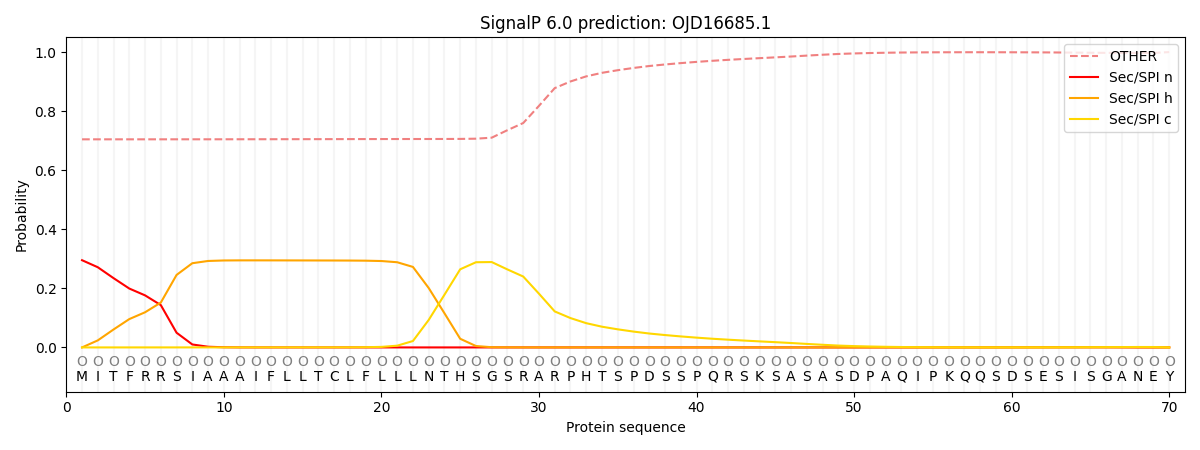
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.719563 | 0.280430 |