You are browsing environment: FUNGIDB
CAZyme Information: OEJ89444.1
You are here: Home > Sequence: OEJ89444.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Hanseniaspora uvarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Saccharomycodaceae; Hanseniaspora; Hanseniaspora uvarum | |||||||||||
| CAZyme ID | OEJ89444.1 | |||||||||||
| CAZy Family | GT32 | |||||||||||
| CAZyme Description | Chitin synthase 1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.16:26 | 2.4.1.16:26 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 862 | 1028 | 5.6e-67 | 0.9938650306748467 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396286 | Chitin_synth_1 | 2.50e-88 | 862 | 1028 | 1 | 163 | Chitin synthase. This region is found commonly in chitin synthases classes I, II and III. Chitin a linear homopolymer of GlcNAc residues, it is an important component of the cell wall of fungi and is synthesized on the cytoplasmic surface of the cell membrane by membrane bound chitin synthases. |
| 133033 | Chitin_synth_C | 2.71e-77 | 858 | 1173 | 1 | 244 | C-terminal domain of Chitin Synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin. Chitin synthase, also called UDP-N-acetyl-D-glucosamine:chitin 4-beta-N-acetylglucosaminyltransferase, catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of GlcNAc residues formed by covalent beta-1,4 linkages. Chitin is an important component of the cell wall of fungi and bacteria and it is synthesized on the cytoplasmic surface of the cell membrane by membrane bound chitin synthases. Studies with fungi have revealed that most of them contain more than one chitin synthase gene. At least five subclasses of chitin synthases have been identified. |
| 396918 | Tyr_Deacylase | 9.47e-57 | 1520 | 1656 | 14 | 143 | D-Tyr-tRNA(Tyr) deacylase. This family comprises of several D-Tyr-tRNA(Tyr) deacylase proteins. Cell growth inhibition by several d-amino acids can be explained by an in vivo production of d-aminoacyl-tRNA molecules. Escherichia coli and yeast cells express an enzyme, d-Tyr-tRNA(Tyr) deacylase, capable of recycling such d-aminoacyl-tRNA molecules into free tRNA and d-amino acid. Accordingly, upon inactivation of the genes of the above deacylases, the toxicity of d-amino acids increases. Orthologues of the deacylase are found in many cells. |
| 238316 | Dtyr_deacylase | 3.49e-50 | 1522 | 1657 | 18 | 145 | D-Tyrosyl-tRNAtyr deacylases; a class of tRNA-dependent hydrolases which are capable of hydrolyzing the ester bond of D-Tyrosyl-tRNA reducing the level of cellular D-Tyrosine while recycling the peptidyl-tRNA; found in bacteria and in eukaryotes but not in archea; beta barrel-like fold structure; forms homodimers in which two surface cavities serve as the active site for tRNA binding |
| 224407 | Dtd | 4.74e-50 | 1520 | 1657 | 16 | 145 | D-Tyr-tRNAtyr deacylase [Translation, ribosomal structure and biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 659 | 1518 | 181 | 1037 | |
| 0.0 | 611 | 1518 | 121 | 993 | |
| 0.0 | 662 | 1518 | 158 | 993 | |
| 5.02e-318 | 635 | 1518 | 247 | 1123 | |
| 1.01e-317 | 661 | 1518 | 234 | 1115 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.23e-25 | 1517 | 1657 | 15 | 147 | c-Myc DNA Unwinding Element Binding Protein [Homo sapiens],2OKV_B c-Myc DNA Unwinding Element Binding Protein [Homo sapiens],2OKV_C c-Myc DNA Unwinding Element Binding Protein [Homo sapiens],2OKV_D c-Myc DNA Unwinding Element Binding Protein [Homo sapiens] |
|
| 2.00e-23 | 1520 | 1659 | 16 | 147 | Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Aquifex aeolicus [Aquifex aeolicus VF5] |
|
| 2.31e-23 | 1520 | 1659 | 26 | 159 | Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNF_B Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNF_C Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNF_D Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNF_E Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNF_F Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNP_A Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNP_B Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNP_C Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNP_D Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNP_E Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3KNP_F Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3KO3_A D-tyrosyl-tRNA(Tyr) deacylase from Plasmodium falciparum incomplex with ADP, obtained through soaking native enzyme crystal with the ATP [Plasmodium falciparum 3D7],3KO3_B D-tyrosyl-tRNA(Tyr) deacylase from Plasmodium falciparum incomplex with ADP, obtained through soaking native enzyme crystal with the ATP [Plasmodium falciparum 3D7],3KO3_C D-tyrosyl-tRNA(Tyr) deacylase from Plasmodium falciparum incomplex with ADP, obtained through soaking native enzyme crystal with the ATP [Plasmodium falciparum 3D7],3KO3_D D-tyrosyl-tRNA(Tyr) deacylase from Plasmodium falciparum incomplex with ADP, obtained through soaking native enzyme crystal with the ATP [Plasmodium falciparum 3D7],3KO3_E D-tyrosyl-tRNA(Tyr) deacylase from Plasmodium falciparum incomplex with ADP, obtained through soaking native enzyme crystal with the ATP [Plasmodium falciparum 3D7],3KO3_F D-tyrosyl-tRNA(Tyr) deacylase from Plasmodium falciparum incomplex with ADP, obtained through soaking native enzyme crystal with the ATP [Plasmodium falciparum 3D7],3KO4_A Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO4_B Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO4_C Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO4_D Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO4_E Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO4_F Crystal structure of D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO5_A D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO5_B D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO5_C D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO5_D D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO5_E D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO5_F D-Tyr-tRNA(Tyr) deacylase from Plasmodium falciparum in complex with ADP [Plasmodium falciparum 3D7],3KO7_A DTD from Plasmodium falciparum in complex with D-Lysine [Plasmodium falciparum 3D7],3KO7_B DTD from Plasmodium falciparum in complex with D-Lysine [Plasmodium falciparum 3D7],3KO7_C DTD from Plasmodium falciparum in complex with D-Lysine [Plasmodium falciparum 3D7],3KO7_D DTD from Plasmodium falciparum in complex with D-Lysine [Plasmodium falciparum 3D7],3KO7_E DTD from Plasmodium falciparum in complex with D-Lysine [Plasmodium falciparum 3D7],3KO7_F DTD from Plasmodium falciparum in complex with D-Lysine [Plasmodium falciparum 3D7],3KO9_A DTD from Plasmodium falciparum in complex with D-Arginine [Plasmodium falciparum 3D7],3KO9_B DTD from Plasmodium falciparum in complex with D-Arginine [Plasmodium falciparum 3D7],3KO9_C DTD from Plasmodium falciparum in complex with D-Arginine [Plasmodium falciparum 3D7],3KO9_D DTD from Plasmodium falciparum in complex with D-Arginine [Plasmodium falciparum 3D7],3KO9_E DTD from Plasmodium falciparum in complex with D-Arginine [Plasmodium falciparum 3D7],3KO9_F DTD from Plasmodium falciparum in complex with D-Arginine [Plasmodium falciparum 3D7],3KOB_A DTD from Plasmodium falciparum in complex with D-Glutamic acid [Plasmodium falciparum 3D7],3KOB_B DTD from Plasmodium falciparum in complex with D-Glutamic acid [Plasmodium falciparum 3D7],3KOB_C DTD from Plasmodium falciparum in complex with D-Glutamic acid [Plasmodium falciparum 3D7],3KOB_D DTD from Plasmodium falciparum in complex with D-Glutamic acid [Plasmodium falciparum 3D7],3KOB_E DTD from Plasmodium falciparum in complex with D-Glutamic acid [Plasmodium falciparum 3D7],3KOB_F DTD from Plasmodium falciparum in complex with D-Glutamic acid [Plasmodium falciparum 3D7],3KOC_A DTD from Plasmodium falciparum in complex with D-Histidine [Plasmodium falciparum 3D7],3KOC_B DTD from Plasmodium falciparum in complex with D-Histidine [Plasmodium falciparum 3D7],3KOC_C DTD from Plasmodium falciparum in complex with D-Histidine [Plasmodium falciparum 3D7],3KOC_D DTD from Plasmodium falciparum in complex with D-Histidine [Plasmodium falciparum 3D7],3KOC_E DTD from Plasmodium falciparum in complex with D-Histidine [Plasmodium falciparum 3D7],3KOC_F DTD from Plasmodium falciparum in complex with D-Histidine [Plasmodium falciparum 3D7],3KOD_A DTD from Plasmodium falciparum in complex with D-Serine [Plasmodium falciparum 3D7],3KOD_B DTD from Plasmodium falciparum in complex with D-Serine [Plasmodium falciparum 3D7],3KOD_C DTD from Plasmodium falciparum in complex with D-Serine [Plasmodium falciparum 3D7],3KOD_D DTD from Plasmodium falciparum in complex with D-Serine [Plasmodium falciparum 3D7],3KOD_E DTD from Plasmodium falciparum in complex with D-Serine [Plasmodium falciparum 3D7],3KOD_F DTD from Plasmodium falciparum in complex with D-Serine [Plasmodium falciparum 3D7],3LMT_A Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMT_B Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMT_C Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMT_D Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMT_E Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMT_F Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_A Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_B Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_C Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_D Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_E Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_F Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_G Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMU_H Crystal structure of DTD from Plasmodium falciparum [Plasmodium falciparum 3D7],3LMV_A D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes [Plasmodium falciparum 3D7],3LMV_B D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes [Plasmodium falciparum 3D7],3LMV_C D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes [Plasmodium falciparum 3D7],3LMV_D D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes [Plasmodium falciparum 3D7],3LMV_E D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes [Plasmodium falciparum 3D7],3LMV_F D-Tyr-tRNA(Tyr) Deacylase from plasmodium falciparum in complex with hepes [Plasmodium falciparum 3D7],4NBI_A D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 1.86 Angstrom resolution [Plasmodium falciparum 3D7],4NBI_B D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 1.86 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_A D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_B D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_C D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_D D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_E D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_F D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_G D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],4NBJ_H D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with D-tyrosyl-3'-aminoadenosine at 2.20 Angstrom resolution [Plasmodium falciparum 3D7],5J61_A D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_B D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_C D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_D D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_E D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_F D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_G D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7],5J61_H D-aminoacyl-tRNA deacylase (DTD) from Plasmodium falciparum in complex with glycyl-3'-aminoadenosine at 2.10 Angstrom resolution [Plasmodium falciparum 3D7] |
|
| 3.15e-18 | 1520 | 1656 | 16 | 143 | Structure of YihZ from Haemophilus influenzae (HI0670), a D-Tyr-tRNA(Tyr) deacylase [Haemophilus influenzae Rd KW20] |
|
| 2.93e-15 | 1520 | 1655 | 16 | 143 | D-Tyr tRNATyr deacylase from Escherichia coli [Escherichia coli],1JKE_B D-Tyr tRNATyr deacylase from Escherichia coli [Escherichia coli],1JKE_C D-Tyr tRNATyr deacylase from Escherichia coli [Escherichia coli],1JKE_D D-Tyr tRNATyr deacylase from Escherichia coli [Escherichia coli] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.42e-317 | 661 | 1518 | 234 | 1115 | Chitin synthase 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CHS1 PE=1 SV=2 |
|
| 1.57e-238 | 790 | 1529 | 244 | 999 | Chitin synthase 2 OS=Candida albicans OX=5476 GN=CHS2 PE=2 SV=2 |
|
| 3.70e-224 | 788 | 1530 | 182 | 917 | Chitin synthase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=chsC PE=3 SV=3 |
|
| 1.74e-216 | 780 | 1518 | 148 | 875 | Chitin synthase 3 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=chs-3 PE=3 SV=2 |
|
| 8.61e-216 | 788 | 1518 | 191 | 908 | Chitin synthase 2 OS=Exophiala dermatitidis OX=5970 GN=CHS2 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000053 | 0.000000 |
TMHMM Annotations download full data without filtering help
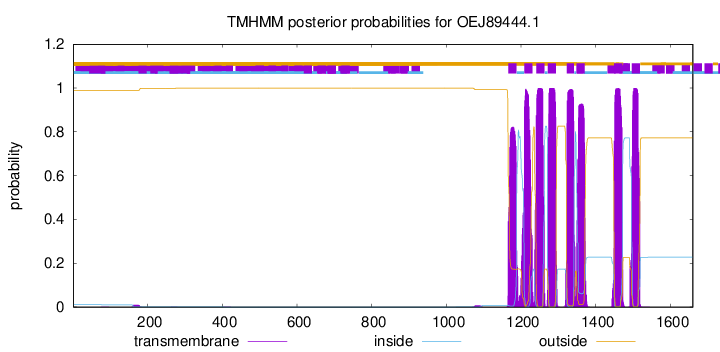
| Start | End |
|---|---|
| 1166 | 1188 |
| 1209 | 1231 |
| 1241 | 1263 |
| 1272 | 1294 |
| 1323 | 1342 |
| 1349 | 1371 |
| 1450 | 1472 |
| 1497 | 1519 |
