You are browsing environment: FUNGIDB
OEJ88841.1
Basic Information
help
Species
Hanseniaspora uvarum
Lineage
Ascomycota; Saccharomycetes; ; Saccharomycodaceae; Hanseniaspora; Hanseniaspora uvarum
CAZyme ID
OEJ88841.1
CAZy Family
GT2
CAZyme Description
Alpha-1,2-mannosyltransferase ALG9
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
625
LPNN01000004|CGC4 72444.52
8.9081
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_HuvarumAWRI3580
4061
N/A
0
4061
Gene Location
Family
Start
End
Evalue
family coverage
GT22
70
500
7.6e-34
0.9897172236503856
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
281842
Glyco_transf_22
1.35e-20
71
500
5
414
Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.27e-40
71
619
19
541
8.20e-40
71
619
19
541
9.24e-39
67
625
10
561
1.74e-38
67
568
15
477
1.74e-38
67
568
15
477
OEJ88841.1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
3.54e-31
67
622
10
548
Alpha-1,2-mannosyltransferase ALG9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ALG9 PE=1 SV=1
more
5.26e-12
89
561
38
500
Alpha-1,2-mannosyltransferase alg9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg9 PE=3 SV=1
more
1.05e-06
86
561
67
509
Dol-P-Man:Man(6)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG9 PE=1 SV=1
more
1.44e-06
70
522
64
504
Alpha-1,2-mannosyltransferase ALG9 OS=Homo sapiens OX=9606 GN=ALG9 PE=1 SV=2
more
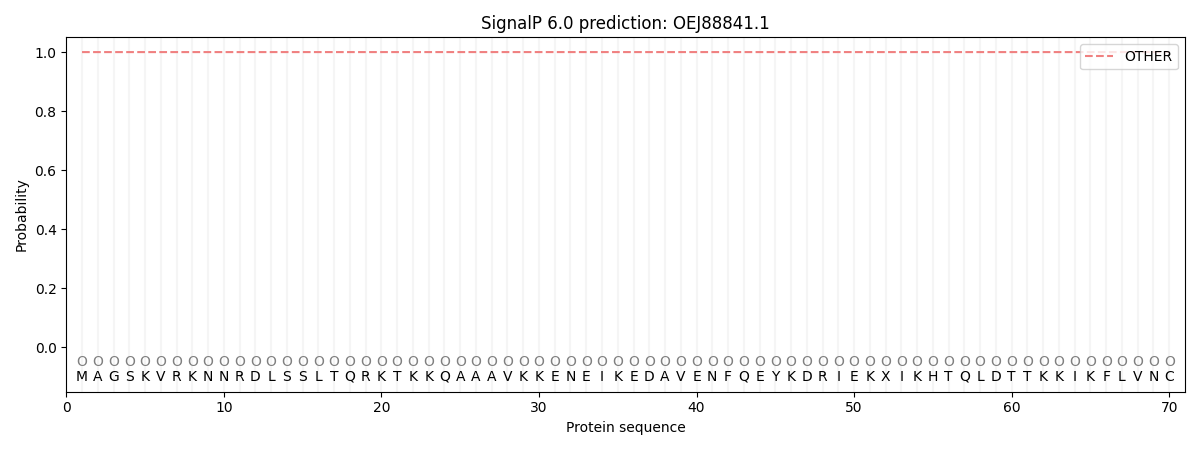
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
1.000052
0.000001
Start
End
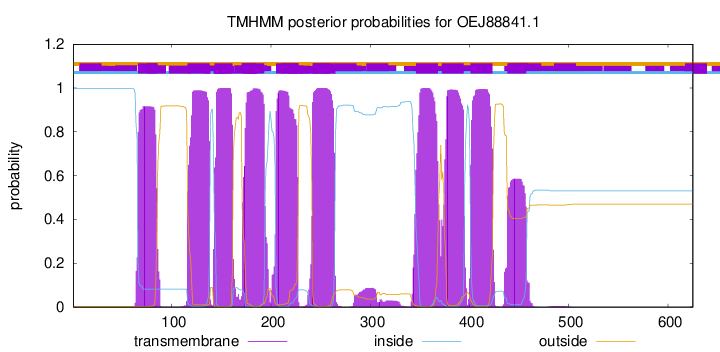
66
85
116
138
143
161
171
193
205
227
242
264
346
368
372
394
401
423
438
457