You are browsing environment: FUNGIDB
CAZyme Information: ODM22257.1
You are here: Home > Sequence: ODM22257.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
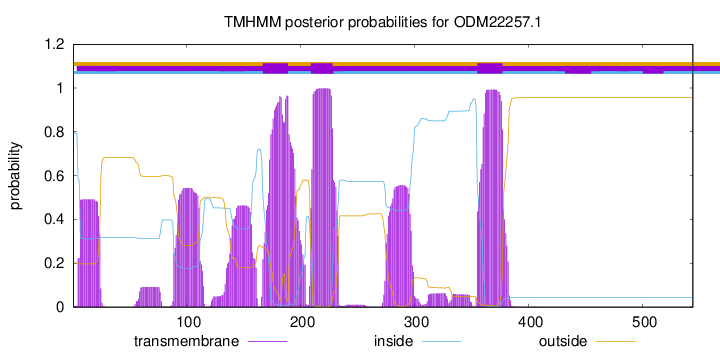
TMHMM annotations
Basic Information help
| Species | Aspergillus cristatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus cristatus | |||||||||||
| CAZyme ID | ODM22257.1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | GPI mannosyltransferase 4 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 5 | 418 | 2.6e-81 | 0.9897172236503856 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 281842 | Glyco_transf_22 | 7.26e-52 | 5 | 417 | 2 | 410 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 544 | 1 | 543 | |
| 2.24e-263 | 1 | 534 | 1 | 533 | |
| 2.24e-263 | 1 | 534 | 1 | 533 | |
| 4.85e-263 | 1 | 534 | 1 | 535 | |
| 4.85e-263 | 1 | 534 | 1 | 535 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.90e-261 | 1 | 534 | 1 | 539 | GPI mannosyltransferase 4 OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=smp3 PE=3 SV=1 |
|
| 1.66e-251 | 1 | 534 | 1 | 543 | GPI mannosyltransferase 4 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=smp3 PE=3 SV=2 |
|
| 9.51e-246 | 1 | 534 | 1 | 538 | GPI mannosyltransferase 4 OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=smp3 PE=3 SV=2 |
|
| 1.00e-186 | 1 | 536 | 1 | 515 | GPI mannosyltransferase 4 OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=SMP3 PE=3 SV=2 |
|
| 3.29e-87 | 10 | 438 | 13 | 404 | GPI mannosyltransferase 4 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=SMP3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
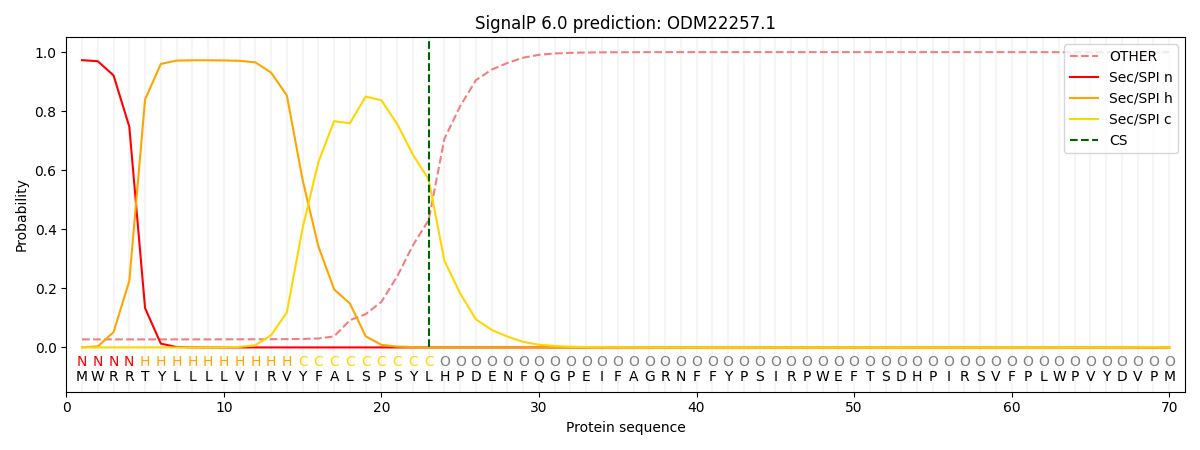
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.034979 | 0.964978 | CS pos: 23-24. Pr: 0.5682 |