You are browsing environment: FUNGIDB
CAZyme Information: ODM20335.1
You are here: Home > Sequence: ODM20335.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aspergillus cristatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Aspergillaceae; Aspergillus; Aspergillus cristatus | |||||||||||
| CAZyme ID | ODM20335.1 | |||||||||||
| CAZy Family | GH55|GH55 | |||||||||||
| CAZyme Description | Mannosyl-oligosaccharide glucosidase [Source:UniProtKB/TrEMBL;Acc:A0A1E3BH76] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.106:25 | 3.2.1.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH63 | 589 | 800 | 2.1e-30 | 0.43157894736842106 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397353 | Glyco_hydro_63 | 0.0 | 304 | 802 | 1 | 494 | Glycosyl hydrolase family 63 C-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the C-terminal catalytic domain. |
| 407154 | Glyco_hydro_63N | 1.23e-124 | 40 | 262 | 1 | 221 | Glycosyl hydrolase family 63 N-terminal domain. This is a family of eukaryotic enzymes belonging to glycosyl hydrolase family 63. They catalyze the specific cleavage of the non-reducing terminal glucose residue from Glc(3)Man(9)GlcNAc(2). Mannosyl oligosaccharide glucosidase EC:3.2.1.106 is the first enzyme in the N-linked oligosaccharide processing pathway. This family represents the N-terminal beta sandwich domain. |
| 225942 | GDB1 | 9.66e-07 | 438 | 750 | 280 | 561 | Glycogen debranching enzyme (alpha-1,6-glucosidase) [Carbohydrate transport and metabolism]. |
| 215307 | PLN02567 | 2.49e-05 | 555 | 797 | 306 | 537 | alpha,alpha-trehalase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 816 | 1 | 816 | |
| 0.0 | 1 | 816 | 1 | 817 | |
| 0.0 | 1 | 816 | 1 | 817 | |
| 0.0 | 1 | 816 | 1 | 829 | |
| 0.0 | 1 | 816 | 1 | 829 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.28e-172 | 30 | 800 | 5 | 800 | Crystal structure of Processing alpha-Glucosidase I [Saccharomyces cerevisiae S288C] |
|
| 2.41e-134 | 40 | 801 | 37 | 776 | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_B Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_C Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus],5MHF_D Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.59e-247 | 25 | 801 | 27 | 805 | Probable mannosyl-oligosaccharide glucosidase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC6G10.09 PE=3 SV=1 |
|
| 9.13e-172 | 19 | 800 | 22 | 830 | Mannosyl-oligosaccharide glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CWH41 PE=1 SV=1 |
|
| 1.64e-134 | 40 | 801 | 92 | 831 | Mannosyl-oligosaccharide glucosidase OS=Mus musculus OX=10090 GN=Mogs PE=1 SV=1 |
|
| 7.86e-131 | 42 | 801 | 94 | 831 | Mannosyl-oligosaccharide glucosidase OS=Rattus norvegicus OX=10116 GN=Mogs PE=1 SV=1 |
|
| 2.04e-127 | 40 | 801 | 94 | 834 | Mannosyl-oligosaccharide glucosidase OS=Homo sapiens OX=9606 GN=MOGS PE=1 SV=5 |
SignalP and Lipop Annotations help
This protein is predicted as SP
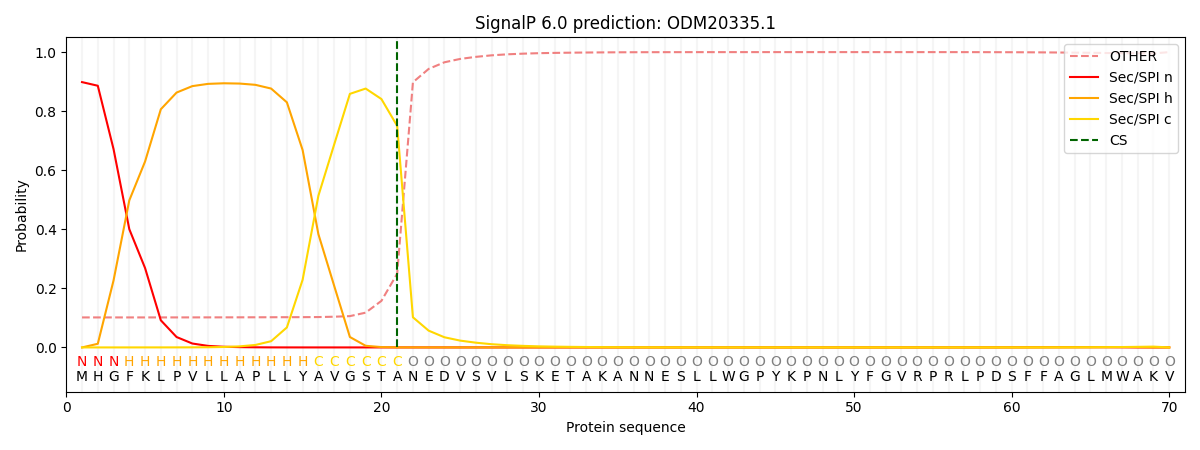
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.122316 | 0.877660 | CS pos: 21-22. Pr: 0.7502 |
