You are browsing environment: FUNGIDB
CAZyme Information: OAT10745.1
You are here: Home > Sequence: OAT10745.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Blastomyces gilchristii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Blastomyces; Blastomyces gilchristii | |||||||||||
| CAZyme ID | OAT10745.1 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | DNA-directed RNA polymerase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA11 | 20 | 205 | 1.1e-62 | 0.9685863874345549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 411414 | MSCRAMM_ClfB | 4.47e-05 | 173 | 316 | 503 | 628 | MSCRAMM family adhesin clumping factor ClfB. Clumping factor B is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.35e-147 | 3 | 338 | 5 | 344 | |
| 7.03e-72 | 12 | 203 | 10 | 210 | |
| 4.56e-71 | 14 | 203 | 14 | 212 | |
| 3.85e-70 | 12 | 210 | 10 | 216 | |
| 3.85e-70 | 12 | 210 | 10 | 216 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.29e-33 | 20 | 204 | 1 | 201 | Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
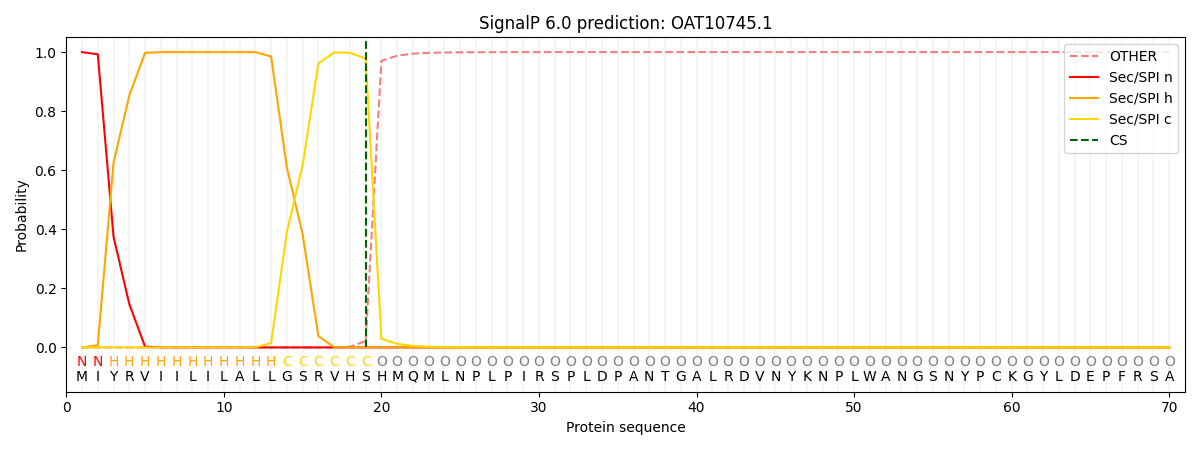
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000220 | 0.999764 | CS pos: 19-20. Pr: 0.9785 |
