You are browsing environment: FUNGIDB
CAZyme Information: OAT08760.1
You are here: Home > Sequence: OAT08760.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
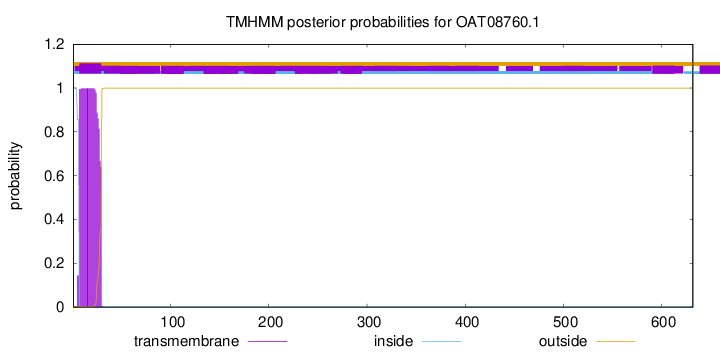
TMHMM annotations
Basic Information help
| Species | Blastomyces gilchristii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Ajellomycetaceae; Blastomyces; Blastomyces gilchristii | |||||||||||
| CAZyme ID | OAT08760.1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | capsule-associated protein CAP1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214773 | CAP10 | 1.09e-15 | 369 | 619 | 28 | 251 | Putative lipopolysaccharide-modifying enzyme. |
| 310354 | Glyco_transf_90 | 6.57e-08 | 373 | 619 | 108 | 321 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 632 | 7 | 624 | |
| 0.0 | 1 | 632 | 1 | 618 | |
| 0.0 | 1 | 632 | 131 | 743 | |
| 3.12e-292 | 91 | 630 | 63 | 598 | |
| 1.89e-281 | 90 | 632 | 52 | 590 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.70e-08 | 373 | 621 | 154 | 379 | Crystal structure of Drosophila Poglut1 (Rumi) complexed with its glycoprotein product (glucosylated EGF repeat) and UDP [Drosophila melanogaster],5F85_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) and UDP [Drosophila melanogaster],5F86_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) [Drosophila melanogaster],5F87_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_B Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_C Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_D Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_E Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_F Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster] |
|
| 3.14e-07 | 350 | 624 | 95 | 345 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.46e-47 | 93 | 627 | 151 | 662 | Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
|
| 2.74e-08 | 373 | 621 | 163 | 388 | O-glucosyltransferase rumi homolog OS=Aedes aegypti OX=7159 GN=AAEL011121 PE=3 SV=1 |
|
| 6.40e-08 | 504 | 621 | 272 | 389 | O-glucosyltransferase rumi homolog OS=Culex quinquefasciatus OX=7176 GN=CPIJ013394 PE=3 SV=1 |
|
| 3.54e-07 | 373 | 621 | 166 | 391 | O-glucosyltransferase rumi OS=Drosophila melanogaster OX=7227 GN=rumi PE=1 SV=1 |
|
| 7.94e-07 | 373 | 621 | 163 | 386 | O-glucosyltransferase rumi homolog OS=Anopheles gambiae OX=7165 GN=AGAP004267 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.952042 | 0.047989 |