You are browsing environment: FUNGIDB
CAZyme Information: OAG25283.1
You are here: Home > Sequence: OAG25283.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alternaria alternata | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Pleosporaceae; Alternaria; Alternaria alternata | |||||||||||
| CAZyme ID | OAG25283.1 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | CBM1 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A177DZY9] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 197593 | fCBD | 5.48e-10 | 23 | 54 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 2.29e-09 | 23 | 50 | 2 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.06e-139 | 1 | 252 | 1 | 249 | |
| 1.27e-119 | 1 | 252 | 1 | 248 | |
| 1.47e-69 | 2 | 249 | 6 | 244 | |
| 1.47e-69 | 2 | 249 | 6 | 244 | |
| 9.21e-69 | 8 | 249 | 12 | 247 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.42e-09 | 18 | 60 | 3 | 45 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 9.65e-08 | 13 | 53 | 368 | 405 | GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8M_B GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8N_A GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872],6Q8N_B GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872] |
|
| 1.25e-06 | 23 | 53 | 7 | 37 | Chain A, ENDOGLUCANASE EG-1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.79e-10 | 3 | 58 | 10 | 65 | Putative endoglucanase type B OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 3.23e-10 | 4 | 57 | 5 | 57 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manF PE=3 SV=1 |
|
| 1.03e-09 | 4 | 58 | 5 | 58 | Mannan endo-1,4-beta-mannosidase F OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=manF PE=3 SV=1 |
|
| 1.08e-09 | 6 | 58 | 8 | 60 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Phanerochaete chrysosporium (strain RP-78 / ATCC MYA-4764 / FGSC 9002) OX=273507 GN=e_gw1.18.61.1 PE=1 SV=1 |
|
| 2.01e-09 | 6 | 60 | 7 | 61 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Sodiomyces alcalophilus OX=398408 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
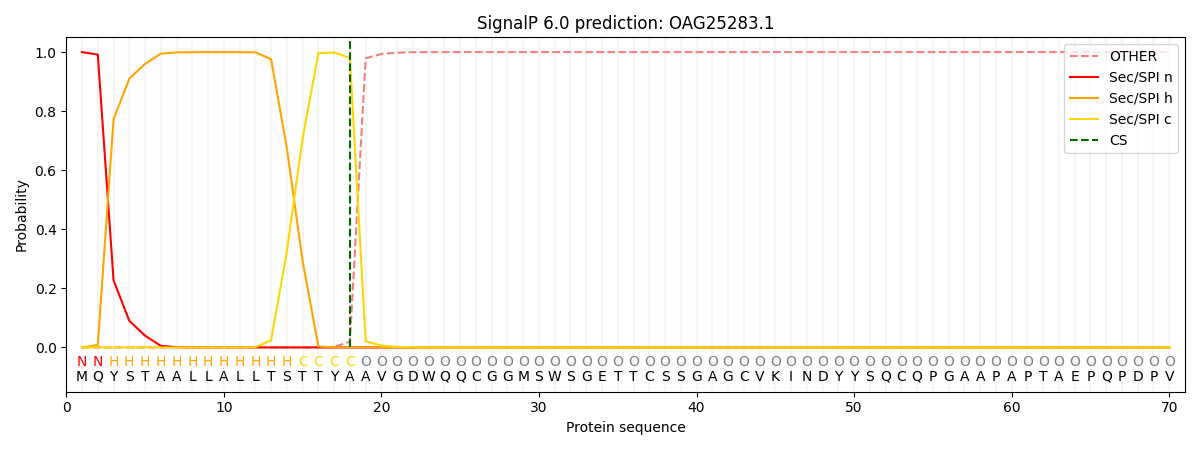
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000212 | 0.999751 | CS pos: 18-19. Pr: 0.9803 |
